#
# Copyright (C) 2024-2025 The University of Sydney, Australia
#
# This program is free software; you can redistribute it and/or modify it under
# the terms of the GNU General Public License, version 2, as published by
# the Free Software Foundation.
#
# This program is distributed in the hope that it will be useful, but WITHOUT
# ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
# FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License
# for more details.
#
# You should have received a copy of the GNU General Public License along
# with this program; if not, write to Free Software Foundation, Inc.,
# 51 Franklin Street, Fifth Floor, Boston, MA 02110-1301, USA.
#
"""A module to generate grids of seafloor age, seafloor spreading rate
and other oceanic data from the `gplately.PlateReconstruction` and
`gplately.plot.PlotTopologies` objects.
Gridding methods in this module have been adapted from Simon Williams'
development repository for an
[auto-age-gridding workflow](https://github.com/siwill22/agegrid-0.1), and are kept
within the `SeafloorGrid` object.
The sample jupyter notebook
[10-SeafloorGrid](https://github.com/GPlates/gplately/blob/master/Notebooks/10-SeafloorGrids.ipynb)
demonstrates how the functionalities within `SeafloorGrid` work. Below you can find
documentation for each of `SeafloorGrid`'s functions.
`SeafloorGrid` Methodology
--------------------------
There are two main steps that `SeafloorGrid` follows to generate grids:
1. Preparation for reconstruction by topologies
2. Reconstruction by topologies
The preparation step involves building a:
* global domain of initial points that populate the seafloor at `max_time`,
* continental mask that separates ocean points from continent regions per timestep, and
* set of points that emerge to the left and right of mid-ocean
ridge segments per timestep, as well as the z-value to allocate to these
points.
First, the global domain of initial points is created using
[stripy's](https://github.com/underworldcode/stripy/blob/master/stripy/spherical_meshes.py#L27)
icosahedral triangulated mesh. The number of points in this mesh can be
controlled using a `refinement_levels` integer (the larger this integer,
the more resolved the continent masks will be).
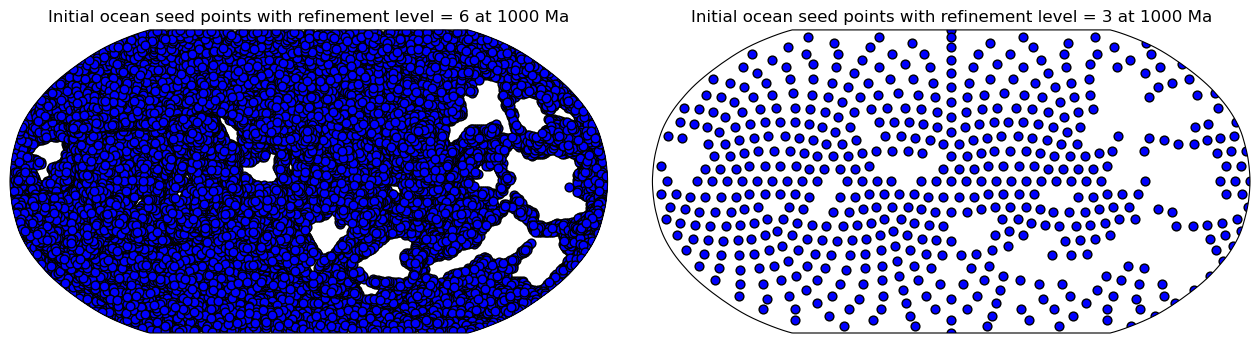
These points are spatially partitioned by plate ID so they can be passed
into a
[point-in-polygon routine](https://gplates.github.io/gplately/oceans.html#gplately.oceans.point_in_polygon_routine).
This identifies points that lie within
continental polygon boundaries and those that are in the ocean. From this,
[continental masks are built](https://gplates.github.io/gplately/oceans.html#gplately.oceans.SeafloorGrid.build_all_continental_masks)
per timestep, and the initial seed points are
allocated ages at the first reconstruction timestep `max_time`. Each point's
initial age is calculated by dividing its proximity to the nearest
MOR segment by half its assumed spreading rate. This spreading rate
(`initial_ocean_mean_spreading_rate`) is assumed to be uniform for all points.
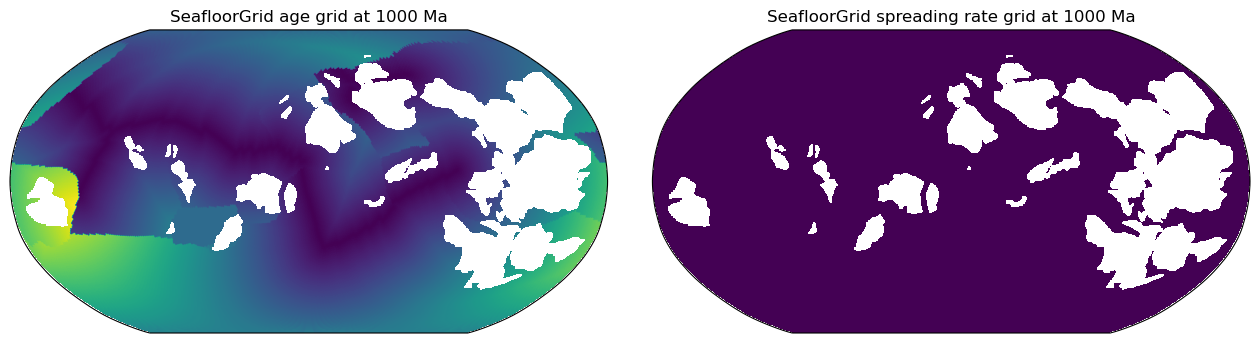
These initial points momentarily fill the global ocean basin, and all have uniform spreading rates.
Thus, the spreading rate grid at `max_time` will be uniformly populated with the `initial_ocean_mean_spreading_rate` (mm/yr).
The age grid at `max_time` will look like a series of smooth, linear age gradients clearly partitioned by
tectonic plates with unique plate IDs:
[Ridge "line" topologies](https://gplates.github.io/gplately/oceans.html#gplately.oceans.SeafloorGrid.build_all_MOR_seedpoints)
are resolved at each reconstruction time step and partitioned
into segments with a valid stage rotation. Each segment is further divided into points
at a specified ridge sampling spacing (`ridge_sampling`). Each point is
ascribed a latitude, longitude, spreading rate and age (from plate reconstruction
model files, as opposed to ages of the initial ocean mesh points), a point index
and the general z-value that will be gridded onto it.

Reconstruction by topologies involves determining which points are active and
inactive (collided with a continent or subducted at a trench) for each reconstruction
time step. This is done using a hidden object in `PlateReconstruction` called
`ReconstructByTopologies`.
If an ocean point with a certain velocity on one plate ID transitions into another
rigid plate ID at another timestep (with another velocity), the velocity difference
between both plates is calculated. The point may have subducted/collided with a continent
if this velocity difference is higher than a specified velocity threshold (which can be
controlled with `subduction_collision_parameters`). To ascertain whether the point
should be deactivated, a displacement test is conducted. If the proximity of the
point's previous time position to the polygon boundary it is approaching is higher than
a set distance threshold, then the point is far enough away from the boundary that it
cannot be subducted or consumed by it, and hence the point is still active. Otherwise,
it is deemed inactive and deleted from the ocean basin mesh.
With each reconstruction time step, points from mid-ocean ridges (which have more
accurate spreading rates and attributed valid times) will spread across the ocean
floor. Eventually, points will be pushed into continental boundaries or subduction
zones, where they are deleted. Ideally, all initial ocean points (from the Stripy
icosahedral mesh) should be deleted over time. However, not all will be deleted -
such points typically reside near continental boundaries. This happens if the
emerged ridge points do not spread far enough to "phase out" these points at
collision regions - likely due to insufficient reconstruction detail. These
undeleted points form artefacts of anomalously high seafloor age that append
over the reconstruction time range.
Once reconstruction by topologies determines the ocean basin snapshot per timestep,
a data frame of all longitudes, latitudes, seafloor ages, spreading rates and any other
attributed z values will be written to a gridding input file per timestep.
Each active longitude, latitude and chosen z value (identified by a gridding input file
column index integer, i.e. `2` is seafloor age) is gridded using nearest-neighbour
interpolation and written to a netCDF4 format.
Classes
-------
* SeafloorGrid
"""
import logging
import math
import multiprocessing
import os
import warnings
from functools import partial
from pathlib import Path
from typing import List, Tuple, Union
import numpy as np
import pandas as pd
import pygplates
from . import grids, tools
from .lib.reconstruct_by_topologies import (
_ContinentCollision,
_DefaultCollision,
_ReconstructByTopologies,
)
from .ptt import continent_contours, separate_ridge_transform_segments
from .tools import _deg2pixels, _pixels2deg
from .utils import seafloor_grid_utils
from .utils.log_utils import get_debug_level
logger = logging.getLogger("gplately")
# JUST FOR PYDOC TO GENERATE THE SAME DOC AS BEFORE
def create_icosahedral_mesh(refinement_levels):
return seafloor_grid_utils.create_icosahedral_mesh(refinement_levels)
def ensure_polygon_geometry(reconstructed_polygons, rotation_model, time):
return seafloor_grid_utils.ensure_polygon_geometry(
reconstructed_polygons, rotation_model, time
)
def point_in_polygon_routine(multi_point, COB_polygons):
return seafloor_grid_utils.point_in_polygon_routine(multi_point, COB_polygons)
create_icosahedral_mesh.__doc__ = seafloor_grid_utils.create_icosahedral_mesh.__doc__
ensure_polygon_geometry.__doc__ = seafloor_grid_utils.ensure_polygon_geometry.__doc__
point_in_polygon_routine.__doc__ = seafloor_grid_utils.point_in_polygon_routine.__doc__
MOR_PKL_FILE_NAME = "MOR_df_{:0.2f}_Ma.pkl"
MOR_GPMLZ_FILE_NAME = "MOR_plus_one_points_{:0.2f}.gpmlz"
SAMPLE_POINTS_GPMLZ_FILE_NAME = "sample_points_{:0.2f}_Ma.gpmlz"
SAMPLE_POINTS_PKL_FILE_NAME = "sample_points_{:0.2f}_Ma.pkl"
[docs]
class SeafloorGrid(object):
"""Generate grids that track data atop global ocean basin points (which emerge from mid-ocean ridges) through geological time."""
[docs]
def __init__(
self,
PlateReconstruction_object,
PlotTopologies_object,
max_time: Union[float, int],
min_time: Union[float, int],
ridge_time_step: Union[float, int],
save_directory: Union[str, Path] = "seafloor-grid-output",
file_collection: str = "",
refinement_levels: int = 5,
ridge_sampling: float = 0.5,
extent: Tuple = (-180, 180, -90, 90),
grid_spacing: float = 0.1,
subduction_collision_parameters=(5.0, 10.0),
initial_ocean_mean_spreading_rate: float = 75.0,
resume_from_checkpoints=False,
zval_names: List[str] = ["SPREADING_RATE"],
continent_mask_filename=None,
use_continent_contouring=False,
):
"""Constructor. Create a :class:`SeafloorGrid` object.
Parameters
----------
PlateReconstruction_object : PlateReconstruction
A :class:`PlateReconstruction` object with a `pygplates.RotationModel`_ and
a `pygplates.FeatureCollection`_ containing topology features.
PlotTopologies_object : PlotTopologies
A :class:`PlotTopologies` object with a continental polygon or COB terrane polygon file to mask grids with.
max_time : float
The maximum time for age gridding.
min_time : float
The minimum time for age gridding.
ridge_time_step : float
The delta time for resolving ridges (and thus age gridding).
save_directory : str, default=None
The top-level directory to save all outputs to.
file_collection : str, default=""
A string to identify the plate model used (will be automated later).
refinement_levels : int, default=5
Control the number of points in the icosahedral mesh (higher integer means higher resolution of continent masks).
ridge_sampling : float, default=0.5
Spatial resolution (in degrees) at which points that emerge from ridges are tessellated.
extent : tuple of 4, default=(-180.,180.,-90.,90.)
A tuple containing the mininum longitude, maximum longitude, minimum latitude and
maximum latitude extents for all masking and final grids.
grid_spacing : float, default=0.1
The degree spacing/interval with which to space grid points across all masking and
final grids. If ``grid_spacing`` is provided, all grids will use it. If not, ``grid_spacing`` defaults to 0.1.
subduction_collision_parameters : len-2 tuple of float, default=(5.0, 10.0)
A 2-tuple of (threshold velocity delta in kms/my, threshold distance to boundary per My in kms/my)
initial_ocean_mean_spreading_rate : float, default=75.
A spreading rate to uniformly allocate to points that define the initial ocean
basin. These points will have inaccurate ages, but most of them will be phased
out after points with plate-model prescribed ages emerge from ridges and spread
to push them towards collision boundaries (where they are deleted).
resume_from_checkpoints : bool, default=False
If set to ``True``, and the gridding preparation stage (continental masking and/or
ridge seed building) is interrupted, SeafloorGrids will resume gridding preparation
from the last successful preparation time.
If set to ``False``, SeafloorGrids will automatically overwrite all files in the
``save_directory`` if re-run after interruption, or normally re-run, thus beginning
gridding preparation from scratch. ``False`` will be useful if data allocated to the
MOR seed points need to be augmented.
zval_names : list of :class:`str`
A list containing string labels for the z values to attribute to points.
Will be used as column headers for z value point dataframes.
continent_mask_filename : str
An optional parameter pointing to the full path to a continental mask for each timestep.
Assuming the time is in the filename, i.e. ``/path/to/continent_mask_0Ma.nc``, it should be
passed as ``/path/to/continent_mask_{}Ma.nc`` with curly brackets. Include decimal formatting if needed.
.. _pygplates.RotationModel: https://www.gplates.org/docs/pygplates/generated/pygplates.rotationmodel
.. _pygplates.Feature: https://www.gplates.org/docs/pygplates/generated/pygplates.feature#pygplates.Feature
.. _pygplates.FeatureCollection: https://www.gplates.org/docs/pygplates/generated/pygplates.featurecollection#pygplates.FeatureCollection
"""
# Provides a rotation model, topology features and reconstruction time for the SeafloorGrid
self.PlateReconstruction_object = PlateReconstruction_object
self.rotation_model = self.PlateReconstruction_object.rotation_model
self.topology_features = self.PlateReconstruction_object.topology_features
self._PlotTopologies_object = PlotTopologies_object
self.topological_model = pygplates.TopologicalModel(
self.topology_features, self.rotation_model
)
self.file_collection = file_collection
if continent_mask_filename:
# Filename for continental masks that the user can provide instead of building it here
self.continent_mask_filepath = continent_mask_filename
self.continent_mask_is_provided = True
else:
self.continent_mask_is_provided = False
self.use_continent_contouring = use_continent_contouring
self._setup_output_paths(save_directory)
# Topological parameters
self.refinement_levels = refinement_levels
self.ridge_sampling = ridge_sampling
self.subduction_collision_parameters = subduction_collision_parameters
self.initial_ocean_mean_spreading_rate = initial_ocean_mean_spreading_rate
# Gridding parameters
self.extent = extent
self._set_grid_resolution(grid_spacing)
self.resume_from_checkpoints = resume_from_checkpoints
# Temporal parameters
self._max_time = max_time
self._min_time = min_time
self._ridge_time_step = ridge_time_step
self._times = np.arange(
self._max_time, self._min_time - 0.1, -self._ridge_time_step
)
# ensure the time for continental masking is consistent.
self._PlotTopologies_object.time = self._max_time
# Essential features and meshes for the SeafloorGrid
self.continental_polygons = ensure_polygon_geometry(
self._PlotTopologies_object.continents, self.rotation_model, self._max_time
)
self._PlotTopologies_object.continents = PlotTopologies_object.continents
self.icosahedral_multi_point = create_icosahedral_mesh(self.refinement_levels)
# Z value parameters
self.zval_names = zval_names
self.default_column_headers = [
"CURRENT_LONGITUDES",
"CURRENT_LATITUDES",
"SEAFLOOR_AGE",
"BIRTH_LAT_SNAPSHOT",
"POINT_ID_SNAPSHOT",
]
self.total_column_headers = self.default_column_headers + self.zval_names
def _map_res_to_node_percentage(self, continent_mask_filename):
"""Determine which percentage to use to scale the continent mask resolution at max time."""
maskY, maskX = grids.read_netcdf_grid(
continent_mask_filename.format(self._max_time)
).shape
mask_deg = _pixels2deg(maskX, self.extent[0], self.extent[1])
if mask_deg <= 0.1:
percentage = 0.1
elif mask_deg <= 0.25:
percentage = 0.3
elif mask_deg <= 0.5:
percentage = 0.5
elif mask_deg < 0.75:
percentage = 0.6
elif mask_deg >= 1:
percentage = 0.75
else:
raise Exception(
"Unable to determine percentage. The code here is suspicious. I am unsure about it."
)
return mask_deg, percentage
def _setup_output_paths(self, save_directory):
"""Create various folders for output files."""
self.save_directory = Path(save_directory)
# zvalue files
self.zvalues_directory = os.path.join(self.save_directory, "zvalues")
Path(self.zvalues_directory).mkdir(parents=True, exist_ok=True)
zvalues_file_basename = "point_data_dataframe_{:0.2f}Ma.npz"
if self.file_collection:
zvalues_file_basename = self.file_collection + "_" + zvalues_file_basename
self.zvalues_file_basepath = os.path.join(
self.zvalues_directory, zvalues_file_basename
)
# middle ocean ridge files
self.mid_ocean_ridges_dir = os.path.join(
self.save_directory, "middle_ocean_ridges"
)
Path(self.mid_ocean_ridges_dir).mkdir(parents=True, exist_ok=True)
if self.file_collection:
self.mid_ocean_ridges_file_path = os.path.join(
self.mid_ocean_ridges_dir,
self.file_collection + "_" + MOR_PKL_FILE_NAME,
)
else:
self.mid_ocean_ridges_file_path = os.path.join(
self.mid_ocean_ridges_dir, MOR_PKL_FILE_NAME
)
# continent mask files
# only generate continent mask files if user does not provide them
if not self.continent_mask_is_provided:
self.continent_mask_directory = os.path.join(
self.save_directory, "continent_mask"
)
Path(self.continent_mask_directory).mkdir(parents=True, exist_ok=True)
if self.use_continent_contouring:
continent_mask_file_basename = (
"continent_mask_ptt_continent_contouring_{:0.2f}Ma.nc"
)
else:
continent_mask_file_basename = "continent_mask_{:0.2f}Ma.nc"
if self.file_collection:
continent_mask_file_basename = (
self.file_collection + "_" + continent_mask_file_basename
)
self.continent_mask_filepath = os.path.join(
self.continent_mask_directory, continent_mask_file_basename
)
# sample points files
self.sample_points_dir = os.path.join(self.save_directory, "sample_points")
Path(self.sample_points_dir).mkdir(parents=True, exist_ok=True)
if self.file_collection:
self.sample_points_file_path = os.path.join(
self.sample_points_dir,
self.file_collection + "_" + SAMPLE_POINTS_PKL_FILE_NAME,
)
else:
self.sample_points_file_path = os.path.join(
self.sample_points_dir, SAMPLE_POINTS_PKL_FILE_NAME
)
# gridding input files
self.gridding_input_directory = os.path.join(
self.save_directory, "gridding_input"
)
Path(self.gridding_input_directory).mkdir(parents=True, exist_ok=True)
gridding_input_basename = "gridding_input_{:0.2f}Ma.npz"
if self.file_collection:
gridding_input_basename = (
self.file_collection + "_" + gridding_input_basename
)
self.gridding_input_filepath = os.path.join(
self.gridding_input_directory, gridding_input_basename
)
def _set_grid_resolution(self, grid_spacing=0.1):
"""Determine the output grid resolution."""
if not grid_spacing:
grid_spacing = 0.1
# A list of degree spacings that allow an even division of the global lat-lon extent.
divisible_degree_spacings = [0.1, 0.25, 0.5, 0.75, 1.0]
# If the provided degree spacing is in the list of permissible spacings, use it
# and prepare the number of pixels in x and y (spacingX and spacingY)
if grid_spacing in divisible_degree_spacings:
self.grid_spacing = grid_spacing
self.spacingX = _deg2pixels(grid_spacing, self.extent[0], self.extent[1])
self.spacingY = _deg2pixels(grid_spacing, self.extent[2], self.extent[3])
# If the provided spacing is >>1 degree, use 1 degree
elif grid_spacing >= divisible_degree_spacings[-1]:
self.grid_spacing = divisible_degree_spacings[-1]
self.spacingX = _deg2pixels(
divisible_degree_spacings[-1], self.extent[0], self.extent[1]
)
self.spacingY = _deg2pixels(
divisible_degree_spacings[-1], self.extent[2], self.extent[3]
)
with warnings.catch_warnings():
warnings.simplefilter("always")
warnings.warn(
f"The provided grid_spacing of {grid_spacing} is quite large. To preserve the grid resolution, a {self.grid_spacing} degree spacing has been employed instead."
)
# If the provided degree spacing is not in the list of permissible spacings, but below
# a degree, find the closest permissible degree spacing. Use this and find
# spacingX and spacingY.
else:
for divisible_degree_spacing in divisible_degree_spacings:
# The tolerance is half the difference between consecutive divisible spacings.
# Max is 1 degree for now - other integers work but may provide too little of a
# grid resolution.
if abs(grid_spacing - divisible_degree_spacing) <= 0.125:
new_deg_res = divisible_degree_spacing
self.grid_spacing = new_deg_res
self.spacingX = _deg2pixels(
new_deg_res, self.extent[0], self.extent[1]
)
self.spacingY = _deg2pixels(
new_deg_res, self.extent[2], self.extent[3]
)
with warnings.catch_warnings():
warnings.simplefilter("always")
warnings.warn(
f"The provided grid_spacing of {grid_spacing} does not cleanly divide into the global extent. A degree spacing of {self.grid_spacing} has been employed instead."
)
# Allow SeafloorGrid time to be updated, and to update the internally-used
# PlotTopologies' time attribute too. If PlotTopologies is used outside the
# object, its `time` attribute is not updated.
@property
def max_time(self):
"""The reconstruction time.
:type: float
"""
return self._max_time
@property
def PlotTopologiesTime(self):
"""The :attr:`PlotTopologies.time` attribute.
:type: float
"""
return self._PlotTopologies_object.time
@max_time.setter
def max_time(self, var):
if var >= 0:
self.update_time(var)
else:
raise ValueError("Enter a valid time >= 0")
[docs]
def update_time(self, max_time: float):
"""Set the new reconstruction time.
Parameters
----------
max_time: float
The new reconstruction time.
"""
self._max_time = float(max_time)
self._PlotTopologies_object.time = float(max_time)
def _collect_point_data_in_dataframe(self, feature_collection, zval_ndarray, time):
"""At a given timestep, create a pandas dataframe holding all attributes of point features.
Rather than store z values as shapefile attributes, store them in a dataframe indexed by feature ID.
"""
return _collect_point_data_in_dataframe(
self.zvalues_file_basepath,
feature_collection,
self.zval_names,
zval_ndarray,
time,
)
def _generate_ocean_points(self):
"""Generate ocean points by using the icosahedral mesh."""
# Ensure COB terranes at max time have reconstruction IDs and valid times
COB_polygons = ensure_polygon_geometry(
self._PlotTopologies_object.continents,
self.rotation_model,
self._max_time,
)
# zval is a binary array encoding whether a point
# coordinate is within a COB terrane polygon or not.
# Use the icosahedral mesh MultiPointOnSphere attribute
_, ocean_basin_point_mesh, zvals = point_in_polygon_routine(
self.icosahedral_multi_point, COB_polygons
)
# Plates to partition with
plate_partitioner = pygplates.PlatePartitioner(
COB_polygons,
self.rotation_model,
)
# Plate partition the ocean basin points
meshnode_feature = pygplates.Feature(
pygplates.FeatureType.create_from_qualified_string("gpml:MeshNode")
)
meshnode_feature.set_geometry(
ocean_basin_point_mesh
# multi_point
)
ocean_basin_meshnode = pygplates.FeatureCollection(meshnode_feature)
paleogeography = plate_partitioner.partition_features(
ocean_basin_meshnode,
partition_return=pygplates.PartitionReturn.separate_partitioned_and_unpartitioned,
properties_to_copy=[pygplates.PropertyName.gpml_shapefile_attributes],
)
return paleogeography[1] # points in oceans
def _get_ocean_points_from_continent_mask(self):
"""Get the ocean points from continent mask grid."""
max_time_cont_mask = grids.Raster(
self.continent_mask_filepath.format(self._max_time)
)
# If the user provides a continental mask filename, we need to downsize the mask
# resolution for when we create the initial ocean mesh. The mesh does not need to be high-res.
# If the input grid is at 0.5 degree uniform spacing, then the input
# grid is 7x more populated than a 6-level stripy icosahedral mesh and
# using this resolution for the initial ocean mesh will dramatically slow down reconstruction by topologies.
# Scale down the resolution based on the input mask resolution
_, percentage = self._map_res_to_node_percentage(self.continent_mask_filepath)
max_time_cont_mask.resize(
int(max_time_cont_mask.shape[0] * percentage),
int(max_time_cont_mask.shape[1] * percentage),
inplace=True,
)
lat = np.linspace(-90, 90, max_time_cont_mask.shape[0])
lon = np.linspace(-180, 180, max_time_cont_mask.shape[1])
llon, llat = np.meshgrid(lon, lat)
mask_inds = np.where(max_time_cont_mask.data.flatten() == 0)
mask_vals = max_time_cont_mask.data.flatten()
mask_lon = llon.flatten()[mask_inds]
mask_lat = llat.flatten()[mask_inds]
ocean_pt_feature = pygplates.Feature()
ocean_pt_feature.set_geometry(
pygplates.MultiPointOnSphere(zip(mask_lat, mask_lon))
)
return [ocean_pt_feature]
[docs]
def create_initial_ocean_seed_points(self):
"""Create the initial ocean basin seed point domain (at ``max_time`` only)
using Stripy's icosahedral triangulation with the specified ``self.refinement_levels``.
The ocean mesh starts off as a global-spanning Stripy icosahedral mesh.
``create_initial_ocean_seed_points`` passes the automatically-resolved-to-current-time
continental polygons from the :attr:`PlotTopologies.continents` attribute
(which can be from a COB terrane file or a continental polygon file) into
Plate Tectonic Tools' point-in-polygon routine. It identifies ocean basin points that lie:
* outside the polygons (for the ocean basin point domain)
* inside the polygons (for the continental mask)
Points from the mesh outside the continental polygons make up the ocean basin seed
point mesh. The masked mesh is outputted as a compressed GPML (GPMLZ) file with
the filename: ``ocean_basin_seed_points_{}Ma.gpmlz`` if a ``save_directory`` is passed.
Otherwise, the mesh is returned as a `pygplates.FeatureCollection`_ object.
.. note::
This point mesh represents ocean basin seafloor that was produced
before :attr:`SeafloorGrid.max_time`, and thus has unknown properties like valid
time and spreading rate. As time passes, the plate reconstruction model sees
points emerging from MORs. These new points spread to occupy the ocean basins,
moving the initial filler points closer to subduction zones and continental
polygons with which they can collide. If a collision is detected, these points are deleted.
Ideally, if a reconstruction tree spans a large time range, **all** initial mesh
points would collide with a continent or be subducted, leaving behind a mesh of
well-defined MOR-emerged ocean basin points that data can be attributed to.
However, some of these initial points situated close to contiental boundaries are
retained through time - these form point artefacts with anomalously high ages. Even
deep-time plate models (e.g. 1 Ga) will have these artefacts - removing them would
require more detail to be added to the reconstruction model.
Returns
-------
ocean_basin_point_mesh : `pygplates.FeatureCollection`_
A `pygplates.FeatureCollection`_ object containing the seed points on the ocean basin.
"""
if (
os.path.isfile(self.continent_mask_filepath.format(self._max_time))
and self.continent_mask_is_provided
):
# If a set of continent masks was passed, we can use max_time's continental
# mask to build the initial profile of seafloor age.
ocean_points = self._get_ocean_points_from_continent_mask()
else:
ocean_points = self._generate_ocean_points()
# Now that we have ocean points...
# Determine age of ocean basin points using their proximity to MOR features
# and an assumed globally-uniform ocean basin mean spreading rate.
# We need resolved topologies at the `max_time` to pass into the proximity function
resolved_topologies = []
shared_boundary_sections = []
pygplates.resolve_topologies(
self.topology_features,
self.rotation_model,
resolved_topologies,
self._max_time,
shared_boundary_sections,
)
pX, pY, pZ = tools.find_distance_to_nearest_ridge(
resolved_topologies,
shared_boundary_sections,
ocean_points,
)
# Divide spreading rate by 2 to use half the mean spreading rate
pAge = np.array(pZ) / (self.initial_ocean_mean_spreading_rate / 2.0)
self._update_current_active_points(
pX,
pY,
pAge + self._max_time,
[0] * len(pX),
[self.initial_ocean_mean_spreading_rate] * len(pX),
)
self.initial_ocean_point_df = self.current_active_points_df
# the code below is for debug purpose only
if get_debug_level() > 100:
initial_ocean_point_features = []
for point in zip(pX, pY, pAge):
point_feature = pygplates.Feature()
point_feature.set_geometry(pygplates.PointOnSphere(point[1], point[0]))
# Add 'time' to the age at the time of computation, to get the valid time in Ma
point_feature.set_valid_time(point[2] + self._max_time, -1)
# For now: custom zvals are added as shapefile attributes - will attempt pandas data frames
# point_feature = set_shapefile_attribute(point_feature, self.initial_ocean_mean_spreading_rate, "SPREADING_RATE") # Seems like static data
initial_ocean_point_features.append(point_feature)
basename = "ocean_basin_seed_points_{}_RLs_{}Ma.gpmlz".format(
self.refinement_levels,
self._max_time,
)
if self.file_collection:
basename = "{}_{}".format(self.file_collection, basename)
initial_ocean_feature_collection = pygplates.FeatureCollection(
initial_ocean_point_features
)
initial_ocean_feature_collection.write(
os.path.join(self.save_directory, basename)
)
# save the zvalue(spreading rate) of the initial ocean points to file "point_data_dataframe_{max_time}Ma.npz"
self._collect_point_data_in_dataframe(
initial_ocean_feature_collection,
np.array(
[self.initial_ocean_mean_spreading_rate] * len(pX)
), # for now, spreading rate is one zvalue for initial ocean points. will other zvalues need to have a generalised workflow?
self._max_time,
)
[docs]
def build_all_MOR_seedpoints(self):
"""Resolve mid-ocean ridges for all times between ``min_time`` and ``max_time``, divide them
into points that make up their shared sub-segments. Rotate these points to the left
and right of the ridge using their stage rotation so that they spread from the ridge.
Z-value allocation to each point is done here. In future, a function (like
the spreading rate function) to calculate general z-data will be an input parameter.
.. note::
If MOR seed point building is interrupted, progress is safeguarded as long as
``resume_from_checkpoints`` is set to ``True``.
This assumes that points spread from ridges symmetrically, with the exception of
large ridge jumps at successive timesteps. Therefore, z-values allocated to ridge-emerging
points will appear symmetrical until changes in spreading ridge geometries create
asymmetries.
In future, this will have a checkpoint save feature so that execution
(which occurs during preparation for ``ReconstructByTopologies`` and can take several hours)
can be safeguarded against run interruptions.
.. seealso::
`Get tessellated points along a mid ocean ridge <https://github.com/siwill22/agegrid-0.1/blob/master/automatic_age_grid_seeding.py#L117>`__.
"""
overwrite = True
if self.resume_from_checkpoints:
overwrite = False
try:
num_cpus = multiprocessing.cpu_count() - 1
except NotImplementedError:
num_cpus = 1
if num_cpus > 1:
with multiprocessing.Pool(num_cpus) as pool:
pool.map(
partial(
_generate_mid_ocean_ridge_points,
delta_time=self._ridge_time_step,
mid_ocean_ridges_file_path=self.mid_ocean_ridges_file_path,
rotation_model=self.rotation_model,
topology_features=self.topology_features,
zvalues_file_basepath=self.zvalues_file_basepath,
zval_names=self.zval_names,
ridge_sampling=self.ridge_sampling,
overwrite=overwrite,
),
self._times[1:],
)
else:
for time in self._times[1:]:
_generate_mid_ocean_ridge_points(
time,
delta_time=self._ridge_time_step,
mid_ocean_ridges_file_path=self.mid_ocean_ridges_file_path,
rotation_model=self.rotation_model,
topology_features=self.topology_features,
zvalues_file_basepath=self.zvalues_file_basepath,
zval_names=self.zval_names,
ridge_sampling=self.ridge_sampling,
overwrite=overwrite,
)
def _create_continental_mask(self, time_array):
"""Create a continental mask for each timestep."""
if time_array[0] != self._max_time:
print(
"Masking interrupted - resuming continental mask building at {} Ma!".format(
time_array[0]
)
)
for time in time_array:
mask_fn = self.continent_mask_filepath.format(time)
if os.path.isfile(mask_fn):
logger.info(
f"Continent mask file exists and will not create again -- {mask_fn}"
)
continue
self._PlotTopologies_object.time = time
geoms = self._PlotTopologies_object.continents
final_grid = grids.rasterise(
geoms,
key=1.0,
shape=(self.spacingY, self.spacingX),
extent=self.extent,
origin="lower",
)
final_grid[np.isnan(final_grid)] = 0.0
grids.write_netcdf_grid(
self.continent_mask_filepath.format(time),
final_grid.astype("i1"),
extent=(-180, 180, -90, 90),
fill_value=None,
)
logger.info(f"Finished building a continental mask at {time} Ma!")
return
def _build_continental_mask(self, time: float, overwrite=False):
"""Create a continental mask for a given time."""
mask_fn = self.continent_mask_filepath.format(time)
if os.path.isfile(mask_fn) and not overwrite:
logger.info(
f"Continent mask file exists and will not create again -- {mask_fn}"
)
return
self._PlotTopologies_object.time = time
final_grid = grids.rasterise(
self._PlotTopologies_object.continents,
key=1.0,
shape=(self.spacingY, self.spacingX),
extent=self.extent,
origin="lower",
)
final_grid[np.isnan(final_grid)] = 0.0
grids.write_netcdf_grid(
self.continent_mask_filepath.format(time),
final_grid.astype("i1"),
extent=(-180, 180, -90, 90),
fill_value=None,
)
logger.info(f"Finished building a continental mask at {time} Ma!")
[docs]
def build_all_continental_masks(self):
"""Create a continental mask to define the ocean basin for all times between ``min_time`` and ``max_time``.
.. note::
Continental masking progress is safeguarded if ever masking is interrupted,
provided that ``resume_from_checkpoints`` is set to ``True``.
The continental masks will be saved to ``continent_mask_{time}Ma.nc`` as compressed netCDF4 files.
"""
if not self.continent_mask_is_provided:
overwrite = True
if self.resume_from_checkpoints:
overwrite = False
if self.use_continent_contouring:
try:
num_cpus = multiprocessing.cpu_count() - 1
except NotImplementedError:
num_cpus = 1
if num_cpus > 1:
with multiprocessing.Pool(num_cpus) as pool:
pool.map(
partial(
_build_continental_mask_with_contouring,
continent_mask_filepath=self.continent_mask_filepath,
rotation_model=self.rotation_model,
continent_features=self._PlotTopologies_object._continents,
overwrite=overwrite,
),
self._times,
)
else:
for time in self._times:
_build_continental_mask_with_contouring(
time,
continent_mask_filepath=self.continent_mask_filepath,
rotation_model=self.rotation_model,
continent_features=self._PlotTopologies_object._continents,
overwrite=overwrite,
)
else:
for time in self._times:
self._build_continental_mask(time, overwrite)
def _extract_zvalues_from_npz_to_ndarray(self, featurecollection, time):
# NPZ file of seedpoint z values that emerged at this time
loaded_npz = np.load(self.zvalues_file_basepath.format(time))
curr_zvalues = np.empty([len(featurecollection), len(self.zval_names)])
for i in range(len(self.zval_names)):
# Account for the 0th index being for point feature IDs
curr_zvalues[:, i] = np.array(loaded_npz["arr_{}".format(i)])
return curr_zvalues
[docs]
def prepare_for_reconstruction_by_topologies(self):
"""Prepare three main auxiliary files for seafloor data gridding:
* Initial ocean seed points (at ``max_time``)
* Continental masks (from ``max_time`` to ``min_time``)
* MOR points (from ``max_time`` to ``min_time``)
Return lists of all attributes for the initial ocean point mesh and
all ridge points for all times in the reconstruction time array.
"""
# INITIAL OCEAN SEED POINT MESH ----------------------------------------------------
self.create_initial_ocean_seed_points()
logger.info("Finished building initial_ocean_seed_points!")
# MOR SEED POINTS AND CONTINENTAL MASKS --------------------------------------------
# The start time for seeding is controlled by whether the overwrite_existing_gridding_inputs
# parameter is set to `True` (in which case the start time is `max_time`). If it is `False`
# and;
# - a run of seeding and continental masking was interrupted, and ridge points were
# checkpointed at n Ma, seeding resumes at n-1 Ma until `min_time` or another interruption
# occurs;
# - seeding was completed but the subsequent gridding input creation was interrupted,
# seeding is assumed completed and skipped. The workflow automatically proceeds to re-gridding.
self.build_all_continental_masks()
self.build_all_MOR_seedpoints()
# load the initial ocean seed points
lons = self.initial_ocean_point_df["lon"].tolist()
lats = self.initial_ocean_point_df["lat"].tolist()
active_points = [
pygplates.PointOnSphere(lat, lon) for lon, lat in zip(lons, lats)
]
appearance_time = self.initial_ocean_point_df["begin_time"].tolist()
birth_lat = lats
prev_lat = lats
prev_lon = lons
zvalues = np.empty((0, len(self.zval_names)))
zvalues = np.concatenate(
(
zvalues,
self.initial_ocean_point_df["SPREADING_RATE"].to_numpy()[..., None],
),
axis=0,
)
for time in self._times[1:]:
# load MOR points for each time step
df = pd.read_pickle(self.mid_ocean_ridges_file_path.format(time))
lons = df["lon"].tolist()
lats = df["lat"].tolist()
active_points += [
pygplates.PointOnSphere(lat, lon) for lon, lat in zip(lons, lats)
]
appearance_time += [time] * len(lons)
birth_lat += lats
prev_lat += lats
prev_lon += lons
zvalues = np.concatenate(
(zvalues, df[self.zval_names[0]].to_numpy()[..., None]), axis=0
)
return active_points, appearance_time, birth_lat, prev_lat, prev_lon, zvalues
def _update_current_active_points(
self, lons, lats, begin_times, end_times, spread_rates, replace=True
):
"""If the `replace` is true, use the new data to replace self.current_active_points_df.
Otherwise, append the new data to the end of self.current_active_points_df"""
data = {
"lon": lons,
"lat": lats,
"begin_time": begin_times,
"end_time": end_times,
"SPREADING_RATE": spread_rates,
}
if replace:
self.current_active_points_df = pd.DataFrame(data=data)
else:
self.current_active_points_df = pd.concat(
[
self.current_active_points_df,
pd.DataFrame(data=data),
],
ignore_index=True,
)
def _update_current_active_points_coordinates(
self, reconstructed_points: List[pygplates.PointOnSphere]
):
"""Update the current active points with the reconstructed coordinates.
The length of `reconstructed_points` must be the same with the length of self.current_active_points_df
"""
assert len(reconstructed_points) == len(self.current_active_points_df)
lons = []
lats = []
begin_times = []
end_times = []
spread_rates = []
for i in range(len(reconstructed_points)):
if reconstructed_points[i]:
lat_lon = reconstructed_points[i].to_lat_lon()
lons.append(lat_lon[1])
lats.append(lat_lon[0])
begin_times.append(self.current_active_points_df.loc[i, "begin_time"])
end_times.append(self.current_active_points_df.loc[i, "end_time"])
spread_rates.append(
self.current_active_points_df.loc[i, "SPREADING_RATE"]
)
self._update_current_active_points(
lons, lats, begin_times, end_times, spread_rates
)
def _remove_continental_points(self, time):
"""remove all the points which are inside continents at `time` from self.current_active_points_df"""
gridZ, gridX, gridY = grids.read_netcdf_grid(
self.continent_mask_filepath.format(time), return_grids=True
)
ni, nj = gridZ.shape
xmin = np.nanmin(gridX)
xmax = np.nanmax(gridX)
ymin = np.nanmin(gridY)
ymax = np.nanmax(gridY)
# TODO
def remove_points_on_continents(row):
i = int(round((ni - 1) * ((row.lat - ymin) / (ymax - ymin))))
j = int(round((nj - 1) * ((row.lon - xmin) / (xmax - xmin))))
i = 0 if i < 0 else i
j = 0 if j < 0 else j
i = ni - 1 if i > ni - 1 else i
j = nj - 1 if j > nj - 1 else j
if gridZ[i, j] > 0:
return False
else:
return True
m = self.current_active_points_df.apply(remove_points_on_continents, axis=1)
self.current_active_points_df = self.current_active_points_df[m]
def _load_middle_ocean_ridge_points(self, time):
"""add middle ocean ridge points at `time` to current_active_points_df"""
df = pd.read_pickle(self.mid_ocean_ridges_file_path.format(time))
self._update_current_active_points(
df["lon"],
df["lat"],
[time] * len(df),
[0] * len(df),
df["SPREADING_RATE"],
replace=False,
)
# obsolete code. keep here for a while. will delete later. -- 2024-05-30
if 0:
fc = pygplates.FeatureCollection(
self.mid_ocean_ridges_file_path.format(time)
)
assert len(self.zval_names) > 0
lons = []
lats = []
begin_times = []
end_times = []
for feature in fc:
lat_lon = feature.get_geometry().to_lat_lon()
valid_time = feature.get_valid_time()
lons.append(lat_lon[1])
lats.append(lat_lon[0])
begin_times.append(valid_time[0])
end_times.append(valid_time[1])
curr_zvalues = self._extract_zvalues_from_npz_to_ndarray(fc, time)
self._update_current_active_points(
lons, lats, begin_times, end_times, curr_zvalues[:, 0], replace=False
)
def _save_gridding_input_data(self, time):
"""save the data into file for creating netcdf file later"""
data_len = len(self.current_active_points_df["lon"])
np.savez_compressed(
self.gridding_input_filepath.format(time),
CURRENT_LONGITUDES=self.current_active_points_df["lon"],
CURRENT_LATITUDES=self.current_active_points_df["lat"],
SEAFLOOR_AGE=self.current_active_points_df["begin_time"] - time,
BIRTH_LAT_SNAPSHOT=[0] * data_len,
POINT_ID_SNAPSHOT=[0] * data_len,
SPREADING_RATE=self.current_active_points_df["SPREADING_RATE"],
)
[docs]
def reconstruct_by_topological_model(self):
"""Use `pygplates.TopologicalModel`_ class to reconstruct seed points.
This method is an alternative to :meth:`reconstruct_by_topologies()` which uses Python code to do the reconstruction.
.. _pygplates.TopologicalModel: https://www.gplates.org/docs/pygplates/generated/pygplates.topologicalmodel
"""
self.create_initial_ocean_seed_points()
logger.info("Finished building initial_ocean_seed_points!")
self.build_all_continental_masks()
self.build_all_MOR_seedpoints()
# not necessary, but put here for readability purpose only
self.current_active_points_df = self.initial_ocean_point_df
time = int(self._max_time)
while True:
self.current_active_points_df.to_pickle(
self.sample_points_file_path.format(time)
)
self._save_gridding_input_data(time)
# save debug file
if get_debug_level() > 100:
_save_seed_points_as_multipoint_coverage(
self.current_active_points_df["lon"],
self.current_active_points_df["lat"],
self.current_active_points_df["begin_time"] - time,
time,
self.sample_points_dir,
)
next_time = time - int(self._ridge_time_step)
if next_time >= int(self._min_time):
points = [
pygplates.PointOnSphere(row.lat, row.lon)
for index, row in self.current_active_points_df.iterrows()
]
# reconstruct_geometry() needs time to be integral value
# https://www.gplates.org/docs/pygplates/generated/pygplates.topologicalmodel#pygplates.TopologicalModel.reconstruct_geometry
reconstructed_time_span = self.topological_model.reconstruct_geometry(
points,
initial_time=time,
youngest_time=next_time,
time_increment=int(self._ridge_time_step),
deactivate_points=pygplates.ReconstructedGeometryTimeSpan.DefaultDeactivatePoints(
threshold_velocity_delta=self.subduction_collision_parameters[0]
/ 10, # cms/yr
threshold_distance_to_boundary=self.subduction_collision_parameters[
1
], # kms/myr
deactivate_points_that_fall_outside_a_network=True,
),
)
reconstructed_points = reconstructed_time_span.get_geometry_points(
next_time, return_inactive_points=True
)
logger.info(
f"Finished topological reconstruction of {len(self.current_active_points_df)} points from {time} to {next_time} Ma."
)
# update the current activate points to prepare for the reconstruction to "next time"
self._update_current_active_points_coordinates(reconstructed_points)
self._remove_continental_points(next_time)
self._load_middle_ocean_ridge_points(next_time)
time = next_time
else:
break
[docs]
def reconstruct_by_topologies(self):
"""Obtain all active ocean seed points which are points that have not been consumed at subduction zones
or have not collided with continental polygons. Active points' latitudes, longitues, seafloor ages, spreading rates and all
other general z-values are saved to a gridding input file (.npz).
"""
logger.info("Preparing all initial files...")
# Obtain all info from the ocean seed points and all MOR points through time, store in
# arrays
(
active_points,
appearance_time,
birth_lat,
prev_lat,
prev_lon,
zvalues,
) = self.prepare_for_reconstruction_by_topologies()
#### Begin reconstruction by topology process:
# Indices for all points (`active_points`) that have existed from `max_time` to `min_time`.
point_id = range(len(active_points))
# Specify the default collision detection region as subduction zones
default_collision = _DefaultCollision(
feature_specific_collision_parameters=[
(
pygplates.FeatureType.gpml_subduction_zone,
self.subduction_collision_parameters,
)
]
)
# In addition to the default subduction detection, also detect continental collisions
collision_spec = _ContinentCollision(
# This filename string should not have a time formatted into it - this is
# taken care of later.
self.continent_mask_filepath,
default_collision,
verbose=False,
)
# Call the reconstruct by topologies object
topology_reconstruction = _ReconstructByTopologies(
self.rotation_model,
self.topology_features,
self._max_time,
self._min_time,
self._ridge_time_step,
active_points,
point_begin_times=appearance_time,
detect_collisions=collision_spec,
)
# Initialise the reconstruction.
topology_reconstruction.begin_reconstruction()
# Loop over the reconstruction times until the end of the reconstruction time span, or until
# all points have entered their valid time range *and* either exited their time range or
# have been deactivated (subducted forward in time or consumed by MOR backward in time).
reconstruction_data = []
while True:
logger.info(
f"Reconstruct by topologies: working on time {topology_reconstruction.get_current_time():0.2f} Ma"
)
# NOTE:
# topology_reconstruction.get_active_current_points() and topology_reconstruction.get_all_current_points()
# are different. The former is a subset of the latter, and it represents all points at the timestep that
# have not collided with a continental or subduction boundary. The remainders in the latter are inactive
# (NoneType) points, which represent the collided points.
# We need to access active point data from topology_reconstruction.get_all_current_points() because it has
# the same length as the list of all initial ocean points and MOR seed points that have ever emerged from
# spreading ridge topologies through `max_time` to `min_time`. Therefore, it protects the time and space
# order in which all MOR points through time were seeded by pyGPlates. At any given timestep, not all these
# points will be active, but their indices are retained. Thus, z value allocation, point latitudes and
# longitudes of active points will be correctly indexed if taking it from
# topology_reconstruction.get_all_current_points().
curr_points = topology_reconstruction.get_active_current_points()
curr_points_including_inactive = (
topology_reconstruction.get_all_current_points()
)
logger.debug(f"the number of current active points is :{len(curr_points)}")
logger.debug(
f"the number of all current points is :{len(curr_points_including_inactive)}"
)
# Collect latitudes and longitudes of currently ACTIVE points in the ocean basin
curr_lat_lon_points = [point.to_lat_lon() for point in curr_points]
if curr_lat_lon_points:
# Get the number of active points at this timestep.
num_current_points = len(curr_points)
# ndarray to fill with active point lats, lons and zvalues
# FOR NOW, the number of gridding input columns is 6:
# 0 = longitude
# 1 = latitude
# 2 = seafloor age
# 3 = birth latitude snapshot
# 4 = point id
# 5 for the default gridding columns above, plus additional zvalues added next
total_number_of_columns = 5 + len(self.zval_names)
gridding_input_data = np.empty(
[num_current_points, total_number_of_columns]
)
# Lons and lats are first and second columns of the ndarray respectively
gridding_input_data[:, 1], gridding_input_data[:, 0] = zip(
*curr_lat_lon_points
)
# NOTE: We need a single index to access data from curr_points_including_inactive AND allocate
# this data to an ndarray with a number of rows equal to num_current_points. This index will
# append +1 after each loop through curr_points_including_inactive.
i = 0
# Get indices and points of all points at `time`, both active and inactive (which are NoneType points that
# have undergone continental collision or subduction at `time`).
for point_index, current_point in enumerate(
curr_points_including_inactive
):
# Look at all active points (these have not collided with a continent or trench)
if current_point is not None:
# Seafloor age
gridding_input_data[i, 2] = (
appearance_time[point_index]
- topology_reconstruction.get_current_time()
)
# Birth latitude (snapshot)
gridding_input_data[i, 3] = birth_lat[point_index]
# Point ID (snapshot)
gridding_input_data[i, 4] = point_id[
point_index
] # The ID of a corresponding point from the original list of all MOR-resolved points
# GENERAL Z-VALUE ALLOCATION
# Z values are 1st index onwards; 0th belongs to the point feature ID (thus +1)
for j in range(len(self.zval_names)):
# Adjusted index - and we have to add j to 5 to account for lat, lon, age, birth lat and point ID,
adjusted_index = 5 + j
# Spreading rate would be first
# Access current zval from the master list of all zvalues for all points that ever existed in time_array
gridding_input_data[i, adjusted_index] = zvalues[
point_index, j
]
# Go to the next active point
i += 1
gridding_input_dictionary = {}
for i in list(range(total_number_of_columns)):
gridding_input_dictionary[self.total_column_headers[i]] = [
list(j) for j in zip(*gridding_input_data)
][i]
data_to_store = [
gridding_input_dictionary[i] for i in gridding_input_dictionary
]
# save debug file
if get_debug_level() > 100:
seafloor_ages = gridding_input_dictionary["SEAFLOOR_AGE"]
logger.debug(
f"The max and min values of seafloor age are: {np.max(seafloor_ages)} - {np.min(seafloor_ages)} ({topology_reconstruction.get_current_time()}Ma)"
)
_save_seed_points_as_multipoint_coverage(
gridding_input_dictionary["CURRENT_LONGITUDES"],
gridding_input_dictionary["CURRENT_LATITUDES"],
gridding_input_dictionary["SEAFLOOR_AGE"],
topology_reconstruction.get_current_time(),
self.sample_points_dir,
)
np.savez_compressed(
self.gridding_input_filepath.format(
topology_reconstruction.get_current_time()
),
*data_to_store,
)
if not topology_reconstruction.reconstruct_to_next_time():
break
logger.info(
f"Reconstruction has been done for {topology_reconstruction.get_current_time()}!"
)
# return reconstruction_data
[docs]
def lat_lon_z_to_netCDF(
self,
zval_name,
time_arr=None,
unmasked=False,
nprocs=1,
):
"""Produce a netCDF4 grid of a z-value identified by its ``zval_name`` for a given time range in ``time_arr``.
Seafloor age can be gridded by passing ``zval_name`` as ``SEAFLOOR_AGE``, and spreading
rate can be gridded with ``SPREADING_RATE``.
Saves all grids to compressed netCDF format in the attributed directory. Grids
can be read into ndarray format using :func:`gplately.read_netcdf_grid()`.
Parameters
----------
zval_name : str
A string identifier for the z-value.
time_arr : list of float, default=None
A time range to turn lons, lats and z-values into netCDF4 grids. If not provided,
``time_arr`` defaults to the full ``time_array`` provided to :class:`SeafloorGrid`.
unmasked : bool, default=False
Save unmasked grids, in addition to masked versions.
nprocs : int, defaullt=1
Number of processes to use for certain operations (requires joblib).
Passed to ``joblib.Parallel``, so -1 means all available processes.
"""
parallel = None
nprocs = int(nprocs)
if nprocs != 1:
try:
from joblib import Parallel
parallel = Parallel(nprocs)
except ImportError:
warnings.warn(
"Could not import joblib; falling back to serial execution"
)
# User can put any time array within SeafloorGrid bounds, but if none
# is provided, it defaults to the attributed time array
if time_arr is None:
time_arr = self._times
if parallel is None:
for time in time_arr:
_lat_lon_z_to_netCDF_time(
time=time,
zval_name=zval_name,
file_collection=self.file_collection,
save_directory=self.save_directory,
total_column_headers=self.total_column_headers,
extent=self.extent,
resX=self.spacingX,
resY=self.spacingY,
unmasked=unmasked,
continent_mask_filename=self.continent_mask_filepath,
gridding_input_filename=self.gridding_input_filepath,
)
else:
from joblib import delayed
parallel(
delayed(_lat_lon_z_to_netCDF_time)(
time=time,
zval_name=zval_name,
file_collection=self.file_collection,
save_directory=self.save_directory,
total_column_headers=self.total_column_headers,
extent=self.extent,
resX=self.spacingX,
resY=self.spacingY,
unmasked=unmasked,
continent_mask_filename=self.continent_mask_filepath,
gridding_input_filename=self.gridding_input_filepath,
)
for time in time_arr
)
[docs]
def save_netcdf_files(
self,
name,
times=None,
unmasked: bool = False,
nprocs: Union[int, None] = None,
):
"""Interpolate the sample points to create regular grids and save as NetCDF files.
Parameters
----------
name: str
The variable name, such as ``SEAFLOOR_AGE`` or ``SPREADING_RATE``.
times: list
A list of times of interest.
unmasked: bool
A flag to indicate if the unmasked grids should be saved.
nprocs: int
The number of processes to use for multiprocessing.
"""
if times is None:
times = self._times
if nprocs is None:
try:
nprocs = multiprocessing.cpu_count() - 1
except NotImplementedError:
nprocs = 1
if nprocs > 1:
with multiprocessing.Pool(nprocs) as pool:
pool.map(
partial(
_save_netcdf_file,
name=name,
file_collection=self.file_collection,
save_directory=self.save_directory,
extent=self.extent,
resX=self.spacingX,
resY=self.spacingY,
unmasked=unmasked,
continent_mask_filename=self.continent_mask_filepath,
sample_points_file_path=self.sample_points_file_path,
),
times,
)
else:
for time in times:
_save_netcdf_file(
time,
name=name,
file_collection=self.file_collection,
save_directory=self.save_directory,
extent=self.extent,
resX=self.spacingX,
resY=self.spacingY,
unmasked=unmasked,
continent_mask_filename=self.continent_mask_filepath,
sample_points_file_path=self.sample_points_file_path,
)
def _save_netcdf_file(
time,
name,
file_collection,
save_directory: Union[str, Path],
extent,
resX,
resY,
continent_mask_filename: str,
sample_points_file_path: str,
unmasked=False,
):
df = pd.read_pickle(sample_points_file_path.format(time))
# drop invalid data
df = df.replace([np.inf, -np.inf], np.nan).dropna()
# Drop duplicate latitudes and longitudes
unique_data = df.drop_duplicates(subset=["lon", "lat"])
# Acquire lons, lats and zvalues for each time
lons = unique_data["lon"].to_list()
lats = unique_data["lat"].to_list()
if name == "SEAFLOOR_AGE":
zdata = (unique_data["begin_time"] - time).to_numpy()
elif name == "SPREADING_RATE":
zdata = unique_data["SPREADING_RATE"].to_numpy()
else:
raise Exception(f"Unknown variable name: {name}")
# Create a regular grid on which to interpolate lats, lons and zdata
extent_globe = extent
grid_lon = np.linspace(extent_globe[0], extent_globe[1], resX)
grid_lat = np.linspace(extent_globe[2], extent_globe[3], resY)
X, Y = np.meshgrid(grid_lon, grid_lat)
# Interpolate lons, lats and zvals over a regular grid using nearest neighbour interpolation
Z = tools.griddata_sphere((lons, lats), zdata, (X, Y), method="nearest")
Z = Z.astype("f4")
unmasked_basename = f"{name}_grid_unmasked_{time:0.2f}_Ma.nc"
grid_basename = f"{name}_grid_{time:0.2f}_Ma.nc"
if file_collection:
unmasked_basename = f"{file_collection}_{unmasked_basename}"
grid_basename = f"{file_collection}_{grid_basename}"
output_dir = os.path.join(save_directory, name)
Path(output_dir).mkdir(parents=True, exist_ok=True)
grid_output_unmasked = os.path.join(output_dir, unmasked_basename)
grid_output = os.path.join(output_dir, grid_basename)
if unmasked:
grids.write_netcdf_grid(
grid_output_unmasked,
Z,
extent=extent,
significant_digits=2,
fill_value=None,
)
# Identify regions in the grid in the continental mask
# We need the continental mask to match the number of nodes
# in the uniform grid defined above. This is important if we
# pass our own continental mask to SeafloorGrid
cont_mask = grids.read_netcdf_grid(
continent_mask_filename.format(time), resize=(resX, resY)
)
# Use the continental mask to mask out continents
Z[cont_mask.astype(bool)] = np.nan
grids.write_netcdf_grid(
grid_output,
Z,
extent=extent,
significant_digits=2,
fill_value=np.nan,
)
logger.info(f"Save {name} netCDF grid at {time:0.2f} Ma completed!")
def _lat_lon_z_to_netCDF_time(
time,
zval_name,
file_collection,
save_directory: Union[str, Path],
total_column_headers,
extent,
resX,
resY,
continent_mask_filename: str,
gridding_input_filename: str,
unmasked=False,
):
# Read the gridding input made by ReconstructByTopologies:
# Use pandas to load in lons, lats and z values from npz files
npz = np.load(gridding_input_filename.format(time))
curr_data = pd.DataFrame.from_dict(
{item: npz[item] for item in npz.files}, orient="columns"
)
curr_data.columns = total_column_headers
# drop invalid data
curr_data = curr_data.replace([np.inf, -np.inf], np.nan)
curr_data = curr_data.dropna(
subset=["CURRENT_LONGITUDES", "CURRENT_LATITUDES", zval_name]
)
if "SEAFLOOR_AGE" == zval_name:
curr_data = curr_data.drop(curr_data[curr_data.SEAFLOOR_AGE > 350].index)
# Drop duplicate latitudes and longitudes
unique_data = curr_data.drop_duplicates(
subset=["CURRENT_LONGITUDES", "CURRENT_LATITUDES"]
)
# Acquire lons, lats and zvalues for each time
lons = unique_data["CURRENT_LONGITUDES"].to_list()
lats = unique_data["CURRENT_LATITUDES"].to_list()
zdata = np.array(unique_data[zval_name].to_list())
# zdata = np.where(zdata > 375, float("nan"), zdata), to deal with vmax in the future
# zdata = np.nan_to_num(zdata)
# Create a regular grid on which to interpolate lats, lons and zdata
extent_globe = extent
grid_lon = np.linspace(extent_globe[0], extent_globe[1], resX)
grid_lat = np.linspace(extent_globe[2], extent_globe[3], resY)
X, Y = np.meshgrid(grid_lon, grid_lat)
# Interpolate lons, lats and zvals over a regular grid using nearest neighbour interpolation
Z = tools.griddata_sphere((lons, lats), zdata, (X, Y), method="nearest")
Z = Z.astype("f4") # float32 precision
unmasked_basename = f"{zval_name}_grid_unmasked_{time:0.2f}Ma.nc"
grid_basename = f"{zval_name}_grid_{time:0.2f}Ma.nc"
if file_collection:
unmasked_basename = f"{file_collection}_{unmasked_basename}"
grid_basename = f"{file_collection}_{grid_basename}"
output_dir = os.path.join(save_directory, zval_name)
Path(output_dir).mkdir(parents=True, exist_ok=True)
grid_output_unmasked = os.path.join(output_dir, unmasked_basename)
grid_output = os.path.join(output_dir, grid_basename)
if unmasked:
grids.write_netcdf_grid(
grid_output_unmasked,
Z,
extent=extent,
significant_digits=2,
fill_value=None,
)
# Identify regions in the grid in the continental mask
# We need the continental mask to match the number of nodes
# in the uniform grid defined above. This is important if we
# pass our own continental mask to SeafloorGrid
cont_mask = grids.read_netcdf_grid(
continent_mask_filename.format(time), resize=(resX, resY)
)
# Use the continental mask to mask out continents
Z[cont_mask.astype(bool)] = np.nan
grids.write_netcdf_grid(
grid_output,
Z,
extent=extent,
significant_digits=2,
fill_value=np.nan,
)
logger.info(f"{zval_name} netCDF grids for {time:0.2f} Ma complete!")
def _save_age_grid_sample_points_to_gpml(
lons, lats, seafloor_ages, paleo_time, output_file_dir
):
"""save sample points to .gpmlz for debug purpose"""
logger.debug(f"saving age grid sample points to gpml file -- {paleo_time} Ma")
features = []
for lon, lat, age in zip(lons, lats, seafloor_ages):
f = pygplates.Feature()
p = pygplates.PointOnSphere(lat, lon)
f.set_geometry(p)
f.set_valid_time(age + paleo_time, paleo_time) # type: ignore
features.append(f)
pygplates.FeatureCollection(features).write(
os.path.join(output_file_dir, SAMPLE_POINTS_GPMLZ_FILE_NAME.format(paleo_time))
)
def _save_seed_points_as_multipoint_coverage(
lons, lats, seafloor_ages, paleo_time, output_file_dir
):
"""save seed points to .gpmlz as multipoint coverage for debug purpose"""
f = pygplates.Feature()
coverage_geometry = pygplates.MultiPointOnSphere(zip(lats, lons))
coverage_scalars = {
pygplates.ScalarType.create_gpml("SeafloorAge"): seafloor_ages,
}
f.set_geometry((coverage_geometry, coverage_scalars))
f.set_valid_time(paleo_time + 0.5, paleo_time - 0.5) # type: ignore
pygplates.FeatureCollection([f]).write(
os.path.join(
output_file_dir, "seed_points_coverage_{:0.2f}_Ma.gpmlz".format(paleo_time)
)
)
def _build_continental_mask_with_contouring(
time: float,
continent_mask_filepath,
rotation_model,
continent_features,
overwrite=False,
):
"""Build the continent mask for a given time using ptt's 'continent contouring' method.
For more information about 'Continent Contouring', visit https://gplates.github.io/gplately/dev-doc/ptt/continent_contours.html.
"""
mask_fn = continent_mask_filepath.format(time)
if os.path.isfile(mask_fn) and not overwrite:
logger.info(
f"Continent mask file exists and will not create again -- {mask_fn}"
)
return
continent_contouring_point_spacing_degrees = 0.25
continent_contouring_area_threshold_square_kms = 0
continent_contouring_area_threshold_steradians = (
continent_contouring_area_threshold_square_kms
/ (pygplates.Earth.mean_radius_in_kms * pygplates.Earth.mean_radius_in_kms)
)
continent_exclusion_area_threshold_square_kms = 800000
continent_exclusion_area_threshold_steradians = (
continent_exclusion_area_threshold_square_kms
/ (pygplates.Earth.mean_radius_in_kms * pygplates.Earth.mean_radius_in_kms)
)
continent_separation_distance_threshold_radians = (
continent_contours.DEFAULT_CONTINENT_SEPARATION_DISTANCE_THRESHOLD_RADIANS
)
def continent_contouring_buffer_and_gap_distance_radians(time, contoured_continent):
# One distance for time interval [1000, 300] and another for time interval [250, 0].
# And linearly interpolate between them over the time interval [300, 250].
pre_pangea_distance_radians = math.radians(2.5) # convert degrees to radians
post_pangea_distance_radians = math.radians(0.0) # convert degrees to radians
if time > 300:
buffer_and_gap_distance_radians = pre_pangea_distance_radians
elif time < 250:
buffer_and_gap_distance_radians = post_pangea_distance_radians
else:
# Linearly interpolate between 250 and 300 Ma.
interp = float(time - 250) / (300 - 250)
buffer_and_gap_distance_radians = (
interp * pre_pangea_distance_radians
+ (1 - interp) * post_pangea_distance_radians
)
# Area of the contoured continent.
area_steradians = contoured_continent.get_area()
# Linearly reduce the buffer/gap distance for contoured continents with area smaller than 1 million km^2.
area_threshold_square_kms = 500000
area_threshold_steradians = area_threshold_square_kms / (
pygplates.Earth.mean_radius_in_kms * pygplates.Earth.mean_radius_in_kms
)
if area_steradians < area_threshold_steradians:
buffer_and_gap_distance_radians *= (
area_steradians / area_threshold_steradians
)
return buffer_and_gap_distance_radians
continent_contouring = continent_contours.ContinentContouring(
rotation_model,
continent_features,
continent_contouring_point_spacing_degrees,
continent_contouring_area_threshold_steradians,
continent_contouring_buffer_and_gap_distance_radians,
continent_exclusion_area_threshold_steradians,
continent_separation_distance_threshold_radians,
)
continent_mask, _ = (
continent_contouring.get_continent_mask_and_contoured_continents(time)
)
grids.write_netcdf_grid(
continent_mask_filepath.format(time),
continent_mask.astype("i1"),
extent=(-180, 180, -90, 90),
fill_value=None,
)
logger.warning(
f"Finished building a continental mask at {time} Ma using ptt's 'Continent Contouring'!"
+ " For more information about 'Continent Contouring', visit https://gplates.github.io/gplately/dev-doc/ptt/continent_contours.html."
)
def _generate_mid_ocean_ridge_points(
time: float,
delta_time: float,
mid_ocean_ridges_file_path: str,
rotation_model,
topology_features,
zvalues_file_basepath,
zval_names,
ridge_sampling,
overwrite=True,
):
"""generate middle ocean ridge seed points at a given time"""
mor_fn = mid_ocean_ridges_file_path.format(time)
if os.path.isfile(mor_fn) and not overwrite:
logger.info(
f"Middle ocean ridge file exists and will not create again.\n{mor_fn}"
)
return
# Points and their z values that emerge from MORs at this time.
shifted_mor_points = []
point_spreading_rates = []
# Resolve topologies to the current time.
resolved_topologies = []
shared_boundary_sections = []
pygplates.resolve_topologies(
topology_features,
rotation_model,
resolved_topologies,
time,
shared_boundary_sections,
)
# pygplates.ResolvedTopologicalSection objects.
for shared_boundary_section in shared_boundary_sections:
if (
shared_boundary_section.get_feature().get_feature_type()
== pygplates.FeatureType.create_gpml("MidOceanRidge")
):
spreading_feature = shared_boundary_section.get_feature()
# Find the stage rotation of the spreading feature in the
# frame of reference of its geometry at the current
# reconstruction time (the MOR is currently actively spreading).
# The stage pole can then be directly geometrically compared
# to the *reconstructed* spreading geometry.
stage_rotation = separate_ridge_transform_segments.get_stage_rotation_for_reconstructed_geometry(
spreading_feature, rotation_model, time
)
if not stage_rotation:
# Skip current feature - it's not a spreading feature.
continue
# Get the stage pole of the stage rotation.
# Note that the stage rotation is already in frame of
# reference of the *reconstructed* geometry at the spreading time.
stage_pole, _ = stage_rotation.get_euler_pole_and_angle()
# One way rotates left and the other right, but don't know
# which - doesn't matter in our example though.
rotate_slightly_off_mor_one_way = pygplates.FiniteRotation(
stage_pole, np.radians(0.01)
)
rotate_slightly_off_mor_opposite_way = (
rotate_slightly_off_mor_one_way.get_inverse()
)
# Iterate over the shared sub-segments.
for shared_sub_segment in shared_boundary_section.get_shared_sub_segments():
# Tessellate MOR section.
mor_points = pygplates.MultiPointOnSphere(
shared_sub_segment.get_resolved_geometry().to_tessellated(
np.radians(ridge_sampling)
)
)
coords = mor_points.to_lat_lon_list()
lats = [i[0] for i in coords]
lons = [i[1] for i in coords]
boundary_feature = shared_boundary_section.get_feature()
left_plate = boundary_feature.get_left_plate(None)
right_plate = boundary_feature.get_right_plate(None)
if left_plate is not None and right_plate is not None:
# Get the spreading rates for all points in this sub segment
(
spreading_rates,
_,
) = tools.calculate_spreading_rates(
time=time,
lons=lons,
lats=lats,
left_plates=[left_plate] * len(lons),
right_plates=[right_plate] * len(lons),
rotation_model=rotation_model,
delta_time=delta_time,
)
else:
spreading_rates = [np.nan] * len(lons)
# Loop through all but the 1st and last points in the current sub segment
for point, rate in zip(
mor_points.get_points()[1:-1],
spreading_rates[1:-1],
):
# Add the point "twice" to the main shifted_mor_points list; once for a L-side
# spread, another for a R-side spread. Then add the same spreading rate twice
# to the list - this therefore assumes spreading rate is symmetric.
shifted_mor_points.append(rotate_slightly_off_mor_one_way * point)
shifted_mor_points.append(
rotate_slightly_off_mor_opposite_way * point
)
point_spreading_rates.extend([rate] * 2)
# save the middle ocean ridges points to .pkl file
lats_lons = [list(point.to_lat_lon()) for point in shifted_mor_points]
df = pd.DataFrame(lats_lons, columns=["lat", "lon"])
df["SPREADING_RATE"] = point_spreading_rates
df.to_pickle(mor_fn)
if get_debug_level() > 100:
# Summarising get_isochrons_for_ridge_snapshot;
# Write out the ridge point born at 'ridge_time' but their position at 'ridge_time - time_step'.
mor_point_features = []
for curr_point in shifted_mor_points:
feature = pygplates.Feature()
feature.set_geometry(curr_point)
feature.set_valid_time(time, -999) # delete - time_step
mor_point_features.append(feature)
mor_points = pygplates.FeatureCollection(mor_point_features)
# Write MOR points at `time` to gpmlz
mor_points.write(
os.path.join(os.path.dirname(mor_fn), MOR_GPMLZ_FILE_NAME.format(time))
)
# write zvalue spreading rates to file point_data_dataframe_{time}Ma.npz
_collect_point_data_in_dataframe(
zvalues_file_basepath, mor_points, zval_names, point_spreading_rates, time
)
logger.info(f"Finished building MOR seedpoints at {time} Ma!")
def _collect_point_data_in_dataframe(
zvalues_file_basepath, feature_collection, zval_names, zval_ndarray, time
):
"""At a given timestep, create a pandas dataframe holding all attributes of point features.
Rather than store z values as shapefile attributes, store them in a dataframe indexed by feature ID.
"""
# Turn the zval_ndarray into a numPy array
zval_ndarray = np.array(zval_ndarray)
# Prepare the zval ndarray (can be of any shape) to be saved with default point data
zvals_to_store = {}
# If only one zvalue (for now, spreading rate)
if zval_ndarray.ndim == 1:
zvals_to_store[zval_names[0]] = zval_ndarray
data_to_store = [zvals_to_store[i] for i in zvals_to_store]
else:
for i in zval_ndarray.shape[1]:
zvals_to_store[zval_names[i]] = [list(j) for j in zip(*zval_ndarray)][i]
data_to_store = [zvals_to_store[i] for i in zvals_to_store]
np.savez_compressed(
zvalues_file_basepath.format(time),
FEATURE_ID=[str(feature.get_feature_id()) for feature in feature_collection],
*data_to_store,
)