Package gplately
Main objects
GPlately's common objects include:
DataServer
The DataServer object automatically downloads and caches files needed for plate reconstructions to a folder in your system.
These plate reconstruction files include rotation models, topology features and static polygons and geometries such as
coastlines, continents and continent-ocean boundaries. Additional data like rasters, grids and feature data can also be installed.
gdownload = gplately.download.DataServer("Muller2019")
# Download plate reconstruction files and geometries from the Müller et al. 2019 model
rotation_model, topology_features, static_polygons = gdownload.get_plate_reconstruction_files()
coastlines, continents, COBs = gdownload.get_topology_geometries()
# Download the Müller et al. 2019 100 Ma age grid
age_grid = gdownload.get_age_grid(time=100)
# Download the ETOPO1 geotiff raster
etopo = gdownload.get_raster("ETOPO1_tif")
DataServer supports the following plate reconstruction file collections which are bundled with the following data:
| Model name string Identifier | Rot. files | Topology features | Static polygons | Coast-lines | Cont-inents | COB | Age grids | SR grids |
|---|---|---|---|---|---|---|---|---|
| Muller2019 | ✅ | ✅ | ✅ | ✅ | ✅ | ✅ | ✅ | ❌ |
| Muller2016 | ✅ | ✅ | ✅ | ✅ | ❌ | ❌ | ✅ | ❌ |
| Merdith2021 | ✅ | ✅ | ✅ | ✅ | ✅ | ❌ | ❌ | ❌ |
| Cao2020 | ✅ | ✅ | ✅ | ✅ | ✅ | ❌ | ❌ | ❌ |
| Clennett2020 | ✅ | ✅ | ✅ | ✅ | ✅ | ❌ | ✅ | ✅ |
| Seton2012 | ✅ | ✅ | ❌ | ✅ | ❌ | ✅ | ✅ | ❌ |
| Matthews2016 | ✅ | ✅ | ✅ | ✅ | ✅ | ❌ | ❌ | ❌ |
| Merdith2017 | ✅ | ✅ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ |
| Li2008 | ✅ | ✅ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ |
| Pehrsson2015 | ✅ | ✅ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ |
| TorsvikCocks2017 | ✅ | ❌ | ❌ | ✅ | ❌ | ❌ | ❌ | ❌ |
| Young2019 | ✅ | ✅ | ✅ | ✅ | ✅ | ❌ | ❌ | ❌ |
| Scotese2008 | ✅ | ✅ | ❌ | ❌ | ✅ | ❌ | ❌ | ❌ |
| Clennett2020_M19 | ✅ | ✅ | ❌ | ✅ | ✅ | ❌ | ❌ | ❌ |
| Clennett2020_S13 | ✅ | ✅ | ❌ | ✅ | ✅ | ❌ | ❌ | ❌ |
| Muller2008 | ✅ | ❌ | ✅ | ❌ | ❌ | ❌ | ❌ | ❌ |
| Muller2022 | ✅ | ✅ | ✅ | ✅ | ✅ | ✅ | ❌ | ❌ |
| Scotese2016 | ✅ | ❌ | ✅ | ✅ | ❌ | ❌ | ❌ | ❌ |
| Shephard2013 | ✅ | ✅ | ✅ | ✅ | ❌ | ❌ | ❌ | ❌ |
PlateReconstruction
The PlateReconstruction object contains tools to reconstruct geological features like tectonic plates and plate boundaries,
and to interrogate plate kinematic data like plate motion velocities, and rates of subduction and seafloor spreading.
# Build a plate reconstruction model using a rotation model, a set of topology features and static polygons
model = gplately.PlateReconstruction(rotation_model, topology_features, static_polygons)
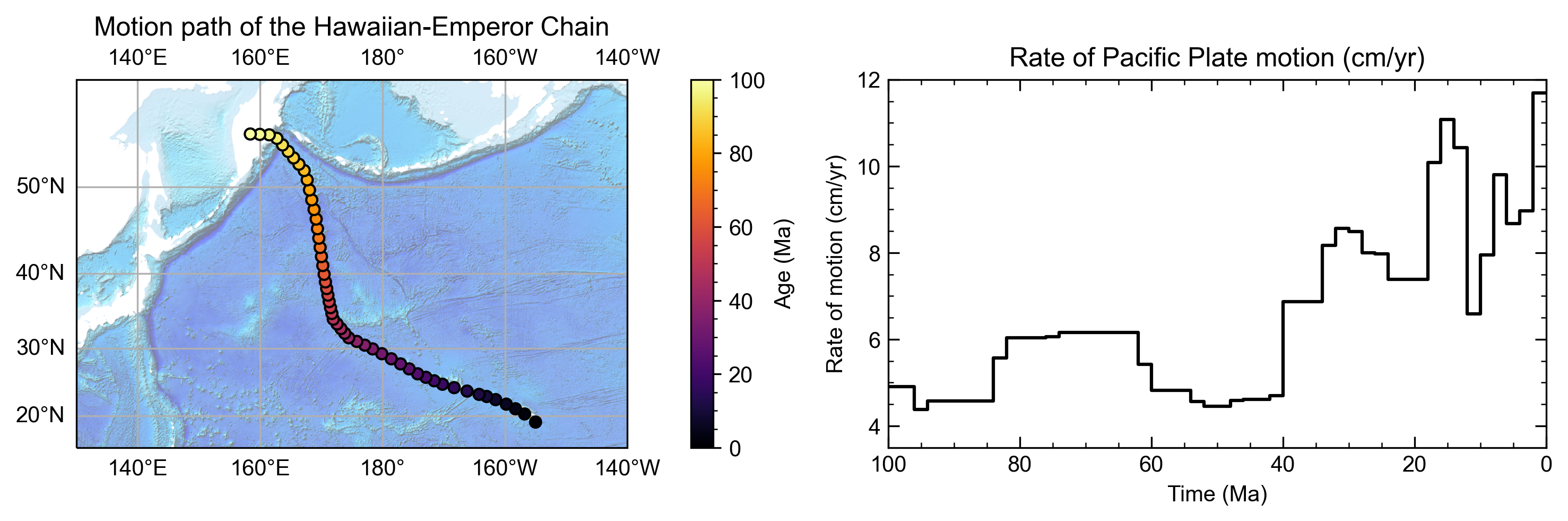
Points
Tools in the Points object track the motion of a point (or group of points) represented by a latitude and longitude
through geologic time. This motion can be visualised using flowlines or motion paths and quantified with point
motion velocities.
# Define some points using their latitude and longitude coordinates so we can track them though time!
pt_lons = np.array([140., 150., 160.])
pt_lats = np.array([-30., -40., -50.])
# Build a Points object from these points
gpts = gplately.Points(model, pt_lons, pt_lats)
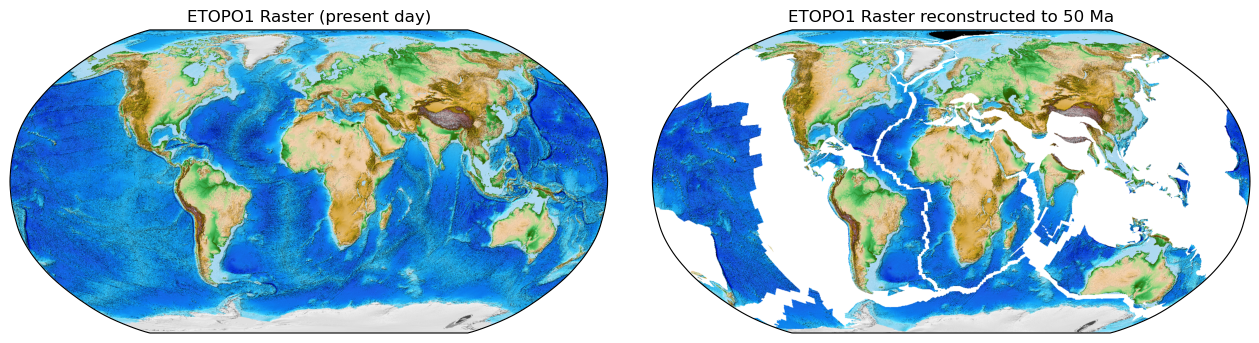
Raster
The Raster object contains tools to work with netCDF4 or MaskedArray gridded data. Grids may be filled,
resized, resampled, and reconstructed back and forwards through geologic time. Other array data can also be
interpolated onto Raster grids.
# Any numpy array can be turned into a Raster object!
raster = gplately.Raster(
plate_reconstruction=model,
data=array,
extent="global", # equivalent to (-180, 180, -90, 90)
origin="lower", # or set extent to (-180, 180, -90, 90)
)
# Reconstruct the raster data to 50 million years ago!
reconstructed_raster = raster.reconstruct(time=50, partitioning_features=continents)
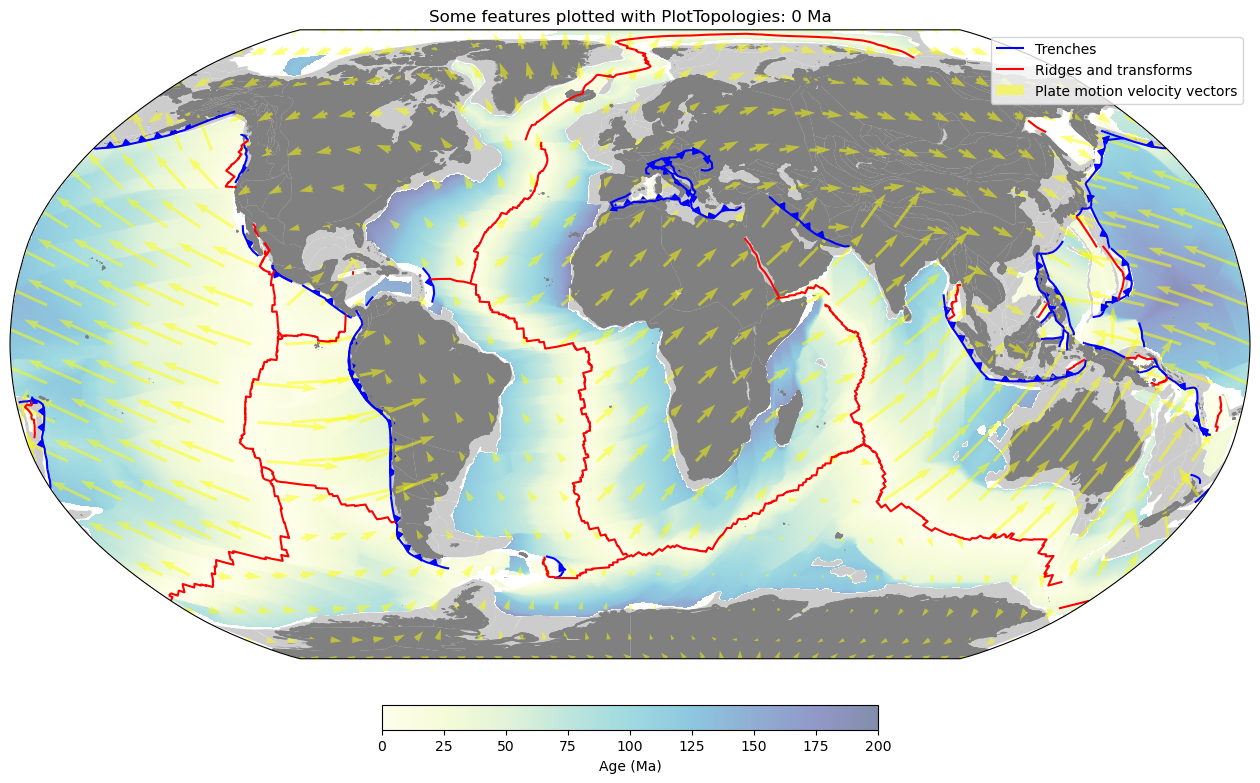
PlotTopologies
PlotTopologies works with the aforementioned PlateReconstruction object to plot
geologic features of different types listed
here, as well as
coastline, continent and continent-ocean boundary geometries reconstructed through time using pyGPlates.
gdownload = gplately.download.DataServer("Muller2019")
# Obtain features for the PlotTopologies object with DataServer
coastlines, continents, COBs = gdownload.get_topology_geometries()
# Call the PlotTopologies object
gplot = gplately.plot.PlotTopologies(
model, # The PlateReconstruction object - it is an input parameter!
time,
coastlines, continents, COBs
)
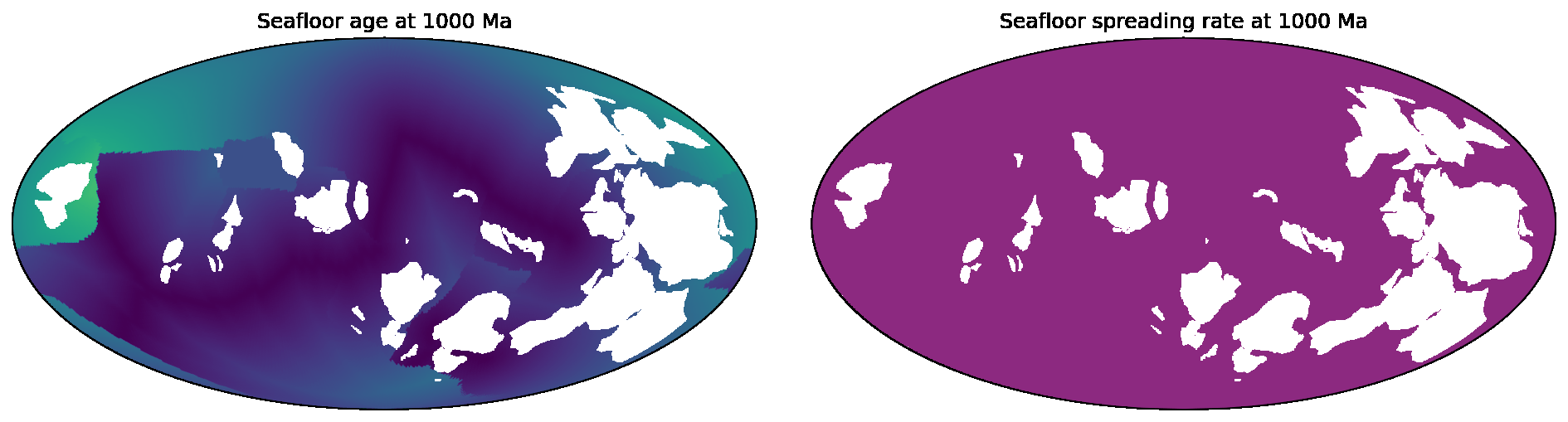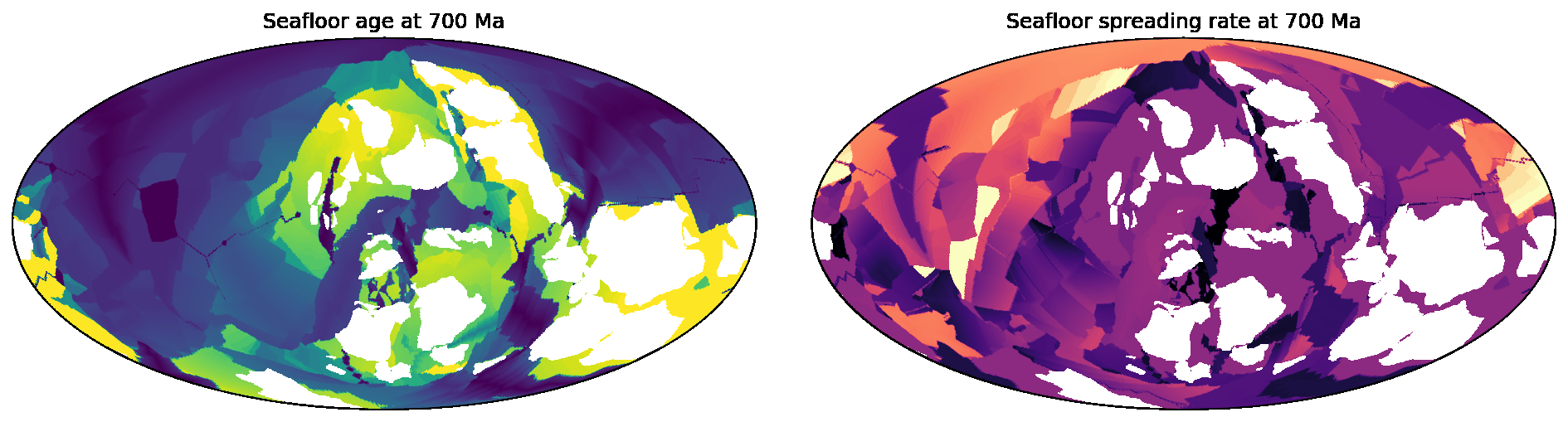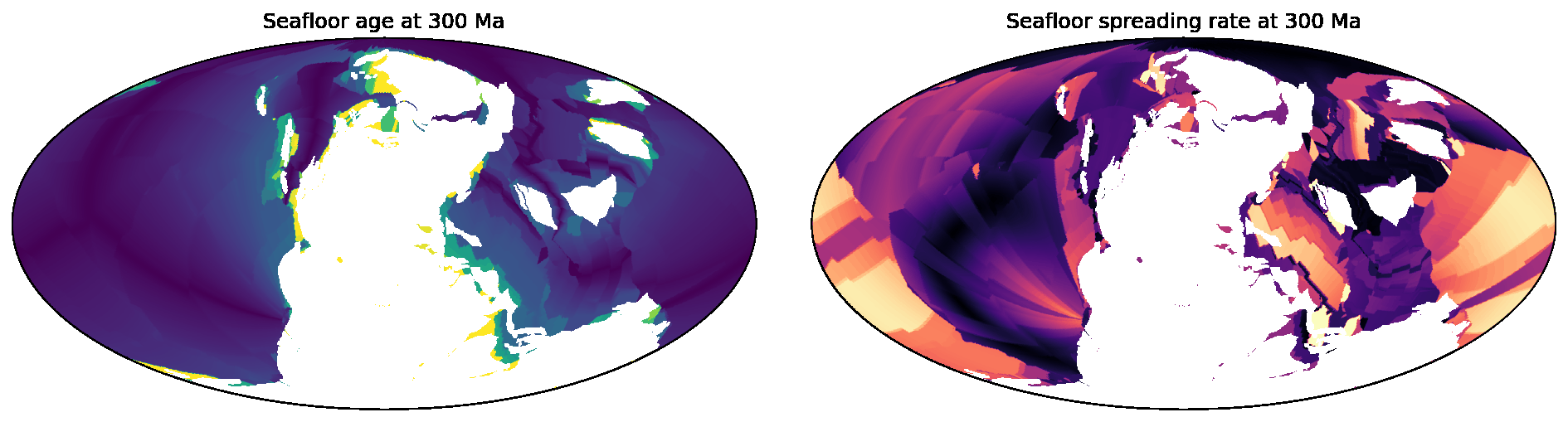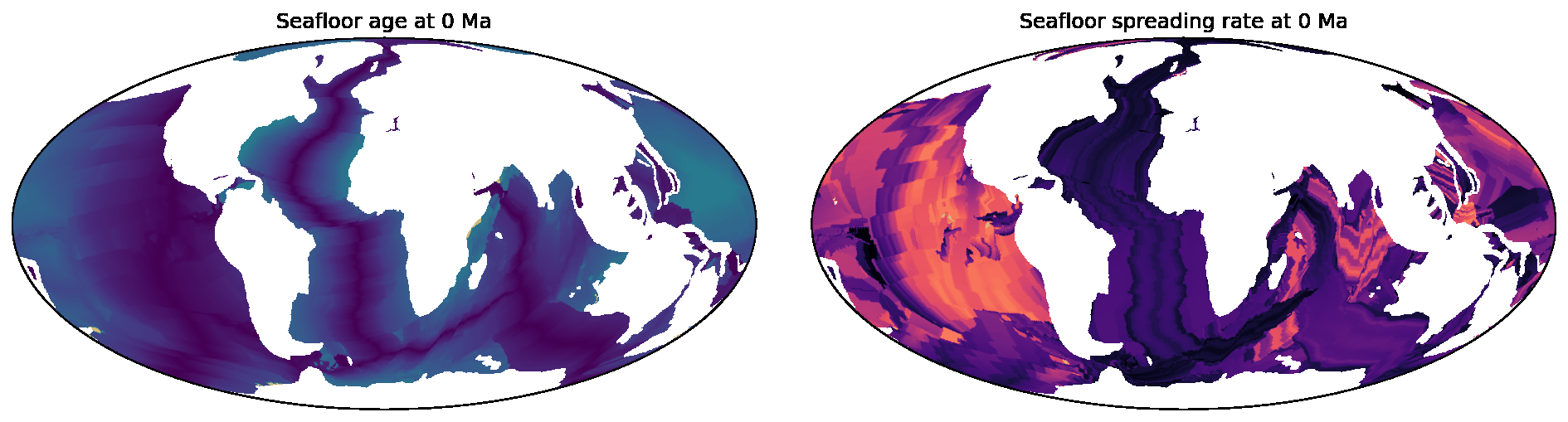
SeafloorGrid
The SeafloorGrid object wraps an automatic workflow to grid seafloor ages and seafloor spreading rates
as encoded by a plate reconstruction model.
10-SeafloorGrids.ipynb is a tutorial notebook that demonstrates
how to set up and use the SeafloorGrid object, and shows a sample set of output grids.
# Set up automatic gridding from 1000Ma to present day
seafloorgrid = gplately.SeafloorGrid(
PlateReconstruction_object = model, #The PlateReconstruction object
PlotTopologies_object = gplot, #The PlotTopologies object
# Time parameters
max_time = 1000, #Ma
min_time = 0, #Ma
)
# Begin automatic gridding!
seafloorgrid.reconstruct_by_topologies()
Notebooks / Examples
- 01 - Getting Started: A brief overview of how to initialise GPlately's main objects
- 02 - Plate Reconstructions: Setting up a
PlateReconstructionobject, reconstructing geological data through time - 03 - Working with Points: Setting up a
Pointsobject, reconstructing seed point locations through time with. This notebook uses point data from the Paleobiology Database (PBDB). - 04 - Velocity Basics: Calculating plate velocities, plotting velocity vector fields
- 05 - Working with Feature Geometries: Processing and plotting assorted polyline, polygon and point data from GPlates 2.3's sample data sets
- 06 - Rasters: Reading, resizing, resampling raster data, and linearly interpolating point data onto raster data
- 07 - Plate Tectonic Stats: Using PlateTectonicTools to calculate and plot subduction zone and ridge data (convergence/spreading velocities, subduction angles, subduction zone and ridge lengths, crustal surface areas produced and subducted etc.)
- 08 - Predicting Slab Flux: Predicting the average slab dip angle of subducting oceanic lithosphere.
- 09 - Motion Paths and Flowlines: Using pyGPlates to create motion paths and flowines of points on a tectonic plate to illustrate the plate's trajectory through geological time.
- 10 - SeafloorGrid: Defines the parameters needed to set up a
SeafloorGridobject, and demonstrates how to produce age and spreading rate grids from a set of plate reconstruction model files.
Expand source code
"""

## Main objects
GPlately's common objects include:
### [DataServer ](https://gplates.github.io/gplately/download.html#gplately.download.DataServer)
The `DataServer` object automatically downloads and caches files needed for plate reconstructions to a folder in your system.
These plate reconstruction files include rotation models, topology features and static polygons and geometries such as
coastlines, continents and continent-ocean boundaries. Additional data like rasters, grids and feature data can also be installed.
```python
gdownload = gplately.download.DataServer("Muller2019")
# Download plate reconstruction files and geometries from the Müller et al. 2019 model
rotation_model, topology_features, static_polygons = gdownload.get_plate_reconstruction_files()
coastlines, continents, COBs = gdownload.get_topology_geometries()
# Download the Müller et al. 2019 100 Ma age grid
age_grid = gdownload.get_age_grid(time=100)
# Download the ETOPO1 geotiff raster
etopo = gdownload.get_raster("ETOPO1_tif")
```
`DataServer` supports the following plate reconstruction file collections which are bundled with the following data:
------------------
| **Model name string Identifier** | **Rot. files** | **Topology features** | **Static polygons** | **Coast-lines** | **Cont-inents** | **COB** | **Age grids** | **SR grids** |
|:--------------------------------:|:--------------:|:---------------------:|:-------------------:|:---------------:|:---------------:|:--------:|:-------------:|:------------:|
| Muller2019 | ✅ | ✅ | ✅ | ✅ | ✅ | ✅ | ✅ | ❌ |
| Muller2016 | ✅ | ✅ | ✅ | ✅ | ❌ | ❌ | ✅ | ❌ |
| Merdith2021 | ✅ | ✅ | ✅ | ✅ | ✅ | ❌ | ❌ | ❌ |
| Cao2020 | ✅ | ✅ | ✅ | ✅ | ✅ | ❌ | ❌ | ❌ |
| Clennett2020 | ✅ | ✅ | ✅ | ✅ | ✅ | ❌ | ✅ | ✅ |
| Seton2012 | ✅ | ✅ | ❌ | ✅ | ❌ | ✅ | ✅ | ❌ |
| Matthews2016 | ✅ | ✅ | ✅ | ✅ | ✅ | ❌ | ❌ | ❌ |
| Merdith2017 | ✅ | ✅ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ |
| Li2008 | ✅ | ✅ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ |
| Pehrsson2015 | ✅ | ✅ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ |
| TorsvikCocks2017 | ✅ | ❌ | ❌ | ✅ | ❌ | ❌ | ❌ | ❌ |
| Young2019 | ✅ | ✅ | ✅ | ✅ | ✅ | ❌ | ❌ | ❌ |
| Scotese2008 | ✅ | ✅ | ❌ | ❌ | ✅ | ❌ | ❌ | ❌ |
| Clennett2020_M19 | ✅ | ✅ | ❌ | ✅ | ✅ | ❌ | ❌ | ❌ |
| Clennett2020_S13 | ✅ | ✅ | ❌ | ✅ | ✅ | ❌ | ❌ | ❌ |
| Muller2008 | ✅ | ❌ | ✅ | ❌ | ❌ | ❌ | ❌ | ❌ |
| Muller2022 | ✅ | ✅ | ✅ | ✅ | ✅ | ✅ | ❌ | ❌ |
| Scotese2016 | ✅ | ❌ | ✅ | ✅ | ❌ | ❌ | ❌ | ❌ |
| Shephard2013 | ✅ | ✅ | ✅ | ✅ | ❌ | ❌ | ❌ | ❌ |
------------------
### [PlateReconstruction](https://gplates.github.io/gplately/reconstruction.html#gplately.reconstruction.PlateReconstruction)
The `PlateReconstruction` object contains tools to reconstruct geological features like tectonic plates and plate boundaries,
and to interrogate plate kinematic data like plate motion velocities, and rates of subduction and seafloor spreading.
```python
# Build a plate reconstruction model using a rotation model, a set of topology features and static polygons
model = gplately.PlateReconstruction(rotation_model, topology_features, static_polygons)
```
### [Points](https://gplates.github.io/gplately/reconstruction.html#gplately.reconstruction.Points)
Tools in the `Points` object track the motion of a point (or group of points) represented by a latitude and longitude
through geologic time. This motion can be visualised using flowlines or motion paths and quantified with point
motion velocities.
```python
# Define some points using their latitude and longitude coordinates so we can track them though time!
pt_lons = np.array([140., 150., 160.])
pt_lats = np.array([-30., -40., -50.])
# Build a Points object from these points
gpts = gplately.Points(model, pt_lons, pt_lats)
```

### [Raster](https://gplates.github.io/gplately/grids.html#gplately.grids.Raster)
The `Raster` object contains tools to work with netCDF4 or MaskedArray gridded data. Grids may be filled,
resized, resampled, and reconstructed back and forwards through geologic time. Other array data can also be
interpolated onto `Raster` grids.
```python
# Any numpy array can be turned into a Raster object!
raster = gplately.Raster(
plate_reconstruction=model,
data=array,
extent="global", # equivalent to (-180, 180, -90, 90)
origin="lower", # or set extent to (-180, 180, -90, 90)
)
# Reconstruct the raster data to 50 million years ago!
reconstructed_raster = raster.reconstruct(time=50, partitioning_features=continents)
```

### [PlotTopologies](https://gplates.github.io/gplately/plot.html#gplately.plot.PlotTopologies)
`PlotTopologies` works with the aforementioned `PlateReconstruction` object to plot
geologic features of different types listed
[here](https://gplates.github.io/gplately/plot.html#gplately.plot.PlotTopologies), as well as
coastline, continent and continent-ocean boundary geometries reconstructed through time using pyGPlates.
```python
gdownload = gplately.download.DataServer("Muller2019")
# Obtain features for the PlotTopologies object with DataServer
coastlines, continents, COBs = gdownload.get_topology_geometries()
# Call the PlotTopologies object
gplot = gplately.plot.PlotTopologies(
model, # The PlateReconstruction object - it is an input parameter!
time,
coastlines, continents, COBs
)
```

### [SeafloorGrid](https://gplates.github.io/gplately/oceans.html#gplately.oceans.SeafloorGrid)
The `SeafloorGrid` object wraps an automatic workflow to grid seafloor ages and seafloor spreading rates
as encoded by a plate reconstruction model.
[10-SeafloorGrids.ipynb](../gplately/Notebooks/10-SeafloorGrids.ipynb) is a tutorial notebook that demonstrates
how to set up and use the `SeafloorGrid` object, and shows a sample set of output grids.
```python
# Set up automatic gridding from 1000Ma to present day
seafloorgrid = gplately.SeafloorGrid(
PlateReconstruction_object = model, #The PlateReconstruction object
PlotTopologies_object = gplot, #The PlotTopologies object
# Time parameters
max_time = 1000, #Ma
min_time = 0, #Ma
)
# Begin automatic gridding!
seafloorgrid.reconstruct_by_topologies()
```

## Notebooks / Examples
- [__01 - Getting Started__](01-GettingStarted.html): A brief overview of how to initialise GPlately's main objects
- [__02 - Plate Reconstructions__](02-PlateReconstructions.html): Setting up a `PlateReconstruction` object, reconstructing geological data through time
- [__03 - Working with Points__](03-WorkingWithPoints.html): Setting up a `Points` object, reconstructing seed point locations through time with. This notebook uses point data from the Paleobiology Database (PBDB).
- [__04 - Velocity Basics__](04-VelocityBasics.html): Calculating plate velocities, plotting velocity vector fields
- [__05 - Working with Feature Geometries__](05-WorkingWithFeatureGeometries.html): Processing and plotting assorted polyline, polygon and point data from [GPlates 2.3's sample data sets](https://www.earthbyte.org/gplates-2-3-software-and-data-sets/)
- [__06 - Rasters__](06-Rasters.html): Reading, resizing, resampling raster data, and linearly interpolating point data onto raster data
- [__07 - Plate Tectonic Stats__](07-WorkingWithPlateTectonicStats.html): Using [PlateTectonicTools](https://github.com/EarthByte/PlateTectonicTools) to calculate and plot subduction zone and ridge data (convergence/spreading velocities, subduction angles, subduction zone and ridge lengths, crustal surface areas produced and subducted etc.)
- [__08 - Predicting Slab Flux__](08-PredictingSlabFlux.html): Predicting the average slab dip angle of subducting oceanic lithosphere.
- [__09 - Motion Paths and Flowlines__](09-CreatingMotionPathsAndFlowlines.html): Using pyGPlates to create motion paths and flowines of points on a tectonic plate to illustrate the plate's trajectory through geological time.
- [__10 - SeafloorGrid__](10-SeafloorGrids.html): Defines the parameters needed to set up a `SeafloorGrid` object, and demonstrates how to produce age and spreading rate grids from a set of plate reconstruction model files.
"""
__version__ = "1.3.0"
try:
import plate_model_manager
except ImportError:
print("The plate_model_manager is not installed, installing it now!")
import subprocess
import sys
subprocess.call([sys.executable, "-m", "pip", "install", "plate-model-manager"])
import plate_model_manager
from . import (
data,
download,
geometry,
gpml,
grids,
oceans,
plot,
ptt,
pygplates,
read_geometries,
reconstruction,
)
from .data import DataCollection
from .download import DataServer
from .grids import Raster
from .oceans import SeafloorGrid
from .plot import PlotTopologies
from .read_geometries import get_geometries, get_valid_geometries
from .reconstruction import (
PlateReconstruction,
Points,
_ContinentCollision,
_DefaultCollision,
_ReconstructByTopologies,
)
from .tools import EARTH_RADIUS
__pdoc__ = {
"data": False,
"_DefaultCollision": False,
"_ContinentCollision": False,
"_ReconstructByTopologies": False,
}
__all__ = [
# Modules
"data",
"download",
"geometry",
"gpml",
"grids",
"oceans",
"plot",
"pygplates",
"read_geometries",
"reconstruction",
"plate_model_manager",
"ptt",
# Classes
"DataCollection",
"DataServer",
"PlateReconstruction",
"PlotTopologies",
"Points",
"Raster",
"SeafloorGrid",
"_ContinentCollision",
"_DefaultCollision",
"_ReconstructByTopologies",
# Functions
"get_geometries",
"get_valid_geometries",
# Constants
"EARTH_RADIUS",
]Sub-modules
gplately.download-
Functions for downloading assorted plate reconstruction data to use with GPlately's main objects. Files are stored in the user's cache and can be …
gplately.examplesgplately.feature_filtergplately.geometry-
Tools for converting PyGPlates or GPlately geometries to Shapely geometries for mapping (and vice versa) …
gplately.gpml-
Tools for manipulating GPML (
.gplately.gpml,.gpmlz) files andFeatureandFeatureCollectionobjects … gplately.grids-
Tools for working with MaskedArray, ndarray and netCDF4 rasters, as well as gridded-data …
gplately.notebooksgplately.oceans-
A module to generate grids of seafloor age, seafloor spreading rate and other oceanic data from the
PlateReconstructionand … gplately.parallel-
Tools to execute routines efficiently by parallelising them over several threads. This uses multiple processing units.
gplately.plot-
Tools for reconstructing and plotting geological features and feature data through time …
gplately.pttgplately.pygplates-
A light wrapping of some
pyGPlatesclasses to keep track of filenames … gplately.read_geometries-
Tools to read geometry data from input files and output them as
Shapelygeometries. These geometries can be plotted directly with GPlately's … gplately.reconstruction-
Tools that wrap up pyGplates and Plate Tectonic Tools functionalities for reconstructing features, working with point data, and calculating plate …
gplately.tools-
A module that offers tools for executing common geological calculations, mathematical conversions and numpy conversions.
Functions
def get_geometries(filename, buffer=None)-
Read a file and return feature geometries.
If
geopandasis available, it will be used to read the file, returning ageopandas.GeoSeries. Ifgeopandasis not found, only shapefiles can be read, and a list ofshapelygeometries will be returned instead of ageopandas.GeoSeries.Parameters
filename:str- Path to the file to be read.
Returns
geometries:listorgeopandas.GeoSeriesshapelygeometries that define the feature geometry held in the shapefile. Can be plotted directly usinggplately.plot.add_geometries.
Expand source code
def get_geometries(filename, buffer=None): """Read a file and return feature geometries. If `geopandas` is available, it will be used to read the file, returning a `geopandas.GeoSeries`. If `geopandas` is not found, only shapefiles can be read, and a list of `shapely` geometries will be returned instead of a `geopandas.GeoSeries`. Parameters ---------- filename : str Path to the file to be read. Returns ------- geometries : list or geopandas.GeoSeries `shapely` geometries that define the feature geometry held in the shapefile. Can be plotted directly using `gplately.plot.add_geometries`. """ if USE_GEOPANDAS: return _get_geometries_geopandas(filename, buffer=buffer) return _get_geometries_cartopy(filename, buffer=buffer) def get_valid_geometries(filename)-
Read a file and return valid feature geometries.
If
geopandasis available, it will be used to read the file, returning ageopandas.GeoSeries. Ifgeopandasis not found, only shapefiles can be read, and a list ofshapelygeometries will be returned instead of ageopandas.GeoSeries.Parameters
filename:str- Path to the file to be read.
Returns
geometries:listorgeopandas.GeoSeries- Valid
shapelygeometries that define the feature geometry held in the shapefile. Can be plotted directly usinggplately.plot.add_geometries.
Expand source code
def get_valid_geometries(filename): """Read a file and return valid feature geometries. If `geopandas` is available, it will be used to read the file, returning a `geopandas.GeoSeries`. If `geopandas` is not found, only shapefiles can be read, and a list of `shapely` geometries will be returned instead of a `geopandas.GeoSeries`. Parameters ---------- filename : str Path to the file to be read. Returns ------- geometries : list or geopandas.GeoSeries Valid `shapely` geometries that define the feature geometry held in the shapefile. Can be plotted directly using `gplately.plot.add_geometries`. """ return get_geometries(filename, buffer=0.0)
Classes
class DataCollection (file_collection)-
GPlately's collection of plate model data is a dictionary where the plate model's identifier string is the key, and values are lists containing any relevant file download links.
Uses a string to identify the needed plate model, taken from
. Expand source code
class DataCollection(object): """GPlately's collection of plate model data is a dictionary where the plate model's identifier string is the key, and values are lists containing any relevant file download links.""" def __init__(self, file_collection): """Uses a string to identify the needed plate model, taken from <gplately.data.DataServer>.""" # Allow strings with capitalisation anywhere. database = [model_name.lower() for model_name in self.plate_reconstruction_files()] if file_collection.lower() not in database: raise ValueError("Enter a valid plate model identifier, e.g. Muller2019, Seton2012, etc.") self.file_collection = file_collection.capitalize() def netcdf4_age_grids(self, time): age_grid_links = { "Muller2019" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Muller_etal_2019_Tectonics/Muller_etal_2019_Agegrids/Muller_etal_2019_Tectonics_v2.0_netCDF/Muller_etal_2019_Tectonics_v2.0_AgeGrid-{}.nc"], "Muller2016" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Muller_etal_2016_AREPS/Muller_etal_2016_AREPS_Agegrids/Muller_etal_2016_AREPS_Agegrids_v1.17/Muller_etal_2016_AREPS_v1.17_netCDF/Muller_etal_2016_AREPS_v1.17_AgeGrid-{}.nc"], "Seton2012" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Seton_etal_2012_ESR/Seton_etal_2012_ESR_Agegrids/netCDF_0-200Ma/agegrid_{}.nc"], "Clennett2020" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Clennett_etal_2020_G3/Clennet_AgeGrids_0.1d_masked/seafloor_age_mask_{}.0Ma.nc"] } links_to_download = _find_needed_collection( self.file_collection, age_grid_links, time) return links_to_download def netcdf4_spreading_rate_grids(self, time): spread_grid_links = { "Clennett2020" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Clennett_etal_2020_G3/Clennett_etal_2020_SpreadRate_Grids/rategrid_final_mask_{}.nc"] } links_to_download = _find_needed_collection( self.file_collection, spread_grid_links, time) return links_to_download def plate_reconstruction_files(self): database = { "Cao2020" : ["https://zenodo.org/record/3854549/files/1000Myr_synthetic_tectonic_reconstructions.zip"], "Muller2019" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Muller_etal_2019_Tectonics/Muller_etal_2019_PlateMotionModel/Muller_etal_2019_PlateMotionModel_v2.0_Tectonics_Updated.zip"], "Muller2016" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Muller_etal_2016_AREPS/Muller_etal_2016_AREPS_Supplement/Muller_etal_2016_AREPS_Supplement_v1.17.zip"], "Clennett2020" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Clennett_etal_2020_G3/Global_Model_WD_Internal_Release_2019_v2_Clennett_NE_Pacific.zip"], "Seton2012" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Seton_etal_2012_ESR.zip"], #"Merdith2021" : ["https://zenodo.org/record/4485738/files/SM2_4485738_V2.zip"], "Merdith2021" : ["https://earthbyte.org/webdav/ftp/Data_Collections/Merdith_etal_2021_ESR/SM2-Merdith_et_al_1_Ga_reconstruction_v1.1.zip"], "Matthews2016" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Matthews_etal_2016_Global_Plate_Model_GPC.zip"], "Merdith2017" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Merdith_etal_2017_GR.zip"], "Li2008" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Li_etal_2008_RodiniaModel.zip"], "Pehrsson2015" : ["https://www.geolsoc.org.uk/~/media/Files/GSL/shared/Sup_pubs/2015/18822_7.zip"], "TorsvikCocks2017" : ["http://www.earthdynamics.org/earthhistory/bookdata/CEED6.zip"], "Young2019" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Young_etal_2018_GeoscienceFrontiers/Young_etal_2018_GeoscienceFrontiers_GPlatesPlateMotionModel.zip"], "Scotese2008" : ["https://static.cambridge.org/content/id/urn:cambridge.org:id:article:S0016756818000110/resource/name/S0016756818000110sup001.zip"], "Golonka2007" : ["https://static.cambridge.org/content/id/urn:cambridge.org:id:article:S0016756818000110/resource/name/S0016756818000110sup001.zip"], "Clennett2020_M2019" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Clennett_etal_2020_G3/Clennett_etal_2020_M2019.zip"], "Clennett2020_S2013" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Clennett_etal_2020_G3/Clennett_etal_2020_S2013.zip"], "Muller2008" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Muller2008/Global_Model_Rigid_Internal_Release_2010.zip"], "Scotese2016" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Scotese2016/PALEOMAP_GlobalPlateModel.zip"], "Shephard2013" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Shephard_etal_2013_ESR.zip"], "Muller2022" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Muller_etal_2022_SE/Muller_etal_2022_SE_1Ga_Opt_PlateMotionModel_v1.1.zip"], "Cao2023" :["https://www.earthbyte.org/webdav/ftp/Data_Collections/Cao_etal_2023/1.8Ga_model_submit.zip"], "Cao2023_Opt" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Cao_etal_2023_Opt/Cao1800Opt.zip"], } return database def plate_model_valid_reconstruction_times(self): database = { "Cao2020" : [0, 1000], "Muller2019" : [0, 250], "Muller2016" : [0, 230], "Clennett2020" : [0, 170], "Seton2012" : [0, 200], "Merdith2021" : [0, 1000], "Matthews2016" : [0, 410], "Merdith2017" : [0, 410], "Li2008" : [0, 410], # "Pehrsson2015" : [25], (First implement continuous rotation) "TorsvikCocks2017" : [0, 410], "Young2019" : [0, 410], "Scotese2008" : [0, 410], "Golonka2007" : [0, 410], "Clennett2020_M2019" : [0, 170], "Clennett2020_S2013" : [0, 170], "Scotese2016" : [0,410], "Shephard2013" : [0,200], "Muller2008" : [0,141], #GPlates static polygons reconstruct to this time "Muller2022" : [0,1000], "Cao2023" : [0,1800], "Cao2023_Opt" : [0,1800], } return database def rotation_strings_to_include(self): strings = [ "Muller2022 1000_0_rotfile_Merdith_et_al_optimised.rot", # For Muller et al. 2022 ] return strings def rotation_strings_to_ignore(self): strings = [ "OLD", "__MACOSX", "DO_NOT", "Blocks_crossing_Poles" ] return strings def dynamic_polygon_strings_to_include(self): strings = [ "plate_boundaries", "PlateBoundaries", "Transform", "Divergence", "Convergence", "Topologies", "Topology", "_PP_", # for Seton 2012 #"ContinentOceanBoundaries", #"Seton_etal_ESR2012_Coastline_2012", "Deforming_Mesh", "Deforming", "Flat_Slabs", "Feature_Geometries", "boundaries", "Clennett_etal_2020_Plates", # For Clennett 2020 (M2019) "Clennett_2020_Plates", # For topologies in Clennett et al 2020 (Pacific) "Clennett_2020_Terranes", # For topologies in Clennett et al 2020 (Pacific) "Angayucham", "Farallon", "Guerrero", "Insular", "Intermontane", "Kula", "North_America", "South_America", "Western_Jurassic", "Clennett_2020_Isochrons", "Clennett_2020_Coastlines", "Clennett_2020_NAm_boundaries", "Shephard_etal_ESR2013_Global_EarthByte_2013", # For Shephard et al. 2013 "1800-1000Ma-plate-boundary_new_valid_time_and_subduction_polarity.gpml", # for Cao2023 ] return strings def dynamic_polygon_strings_to_ignore(self): strings = [ "OLD", "__MACOSX", "DO_NOT", "9_Point", # Muller et al 2019 "9_Point_Density", # Clennett et al 2020 "Density", # Clennett et al 2020 "Inactive_Meshes_and_Topologies", # Clennett et al 2020 "ContinentOceanBoundaries", # Seton 2012 "Seton_etal_ESR2012_Coastline_2012", # Seton 2012 "PALEOMAP_PoliticalBoundaries", # Scotese 2016 "SimplifiedFiles", # Muller et al. 2019 (updated) "1000-410_poles", # Merdith ] return strings def static_polygon_strings_to_include(self): strings = [ "StaticPolygon", "StaticPolygons", "Static_Polygon", "StaticPlatePolygons_", "RodiniaBlocks_WithPlateIDColumnAndIDs", # "PlatePolygons.shp", "CEED6_TERRANES.shp", "CEED6_MICROCONTINENTS.shp", "CEED6_LAND.gpml", "Scotese_2008_PresentDay_ContinentalPolygons", # Scotese 2008 "Golonka_2007_PresentDay_ContinentalPolygons.shp", # Golonka 2007 "PALEOMAP_PlatePolygons.gpml", # For Scotese 2016 ] return strings def static_polygon_strings_to_ignore(self): strings = [ "DO_NOT", "OLD", "__MACOSX", "Global_Model_WD_Internal_Release_2019_v2_Clennett_NE_Pacific/StaticGeometries/StaticPolygons/Global_EarthByte_GPlates_PresentDay_StaticPlatePolygons.shp" # Clennett 2020 ] return strings def topology_geometries(self): database = { "Cao2020" : ["https://zenodo.org/record/3854549/files/1000Myr_synthetic_tectonic_reconstructions.zip"], "Muller2019" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Muller_etal_2019_Tectonics/Muller_etal_2019_PlateMotionModel/Muller_etal_2019_PlateMotionModel_v2.0_Tectonics_Updated.zip"], "Muller2016" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Muller_etal_2016_AREPS/Muller_etal_2016_AREPS_Supplement/Muller_etal_2016_AREPS_Supplement_v1.17.zip"], "Clennett2020" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Clennett_etal_2020_G3/Global_Model_WD_Internal_Release_2019_v2_Clennett_NE_Pacific.zip"], "Seton2012" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Seton_etal_2012_ESR.zip"], #"Merdith2021" : ["https://zenodo.org/record/4485738/files/SM2_4485738_V2.zip"], "Merdith2021" : ["https://earthbyte.org/webdav/ftp/Data_Collections/Merdith_etal_2021_ESR/SM2-Merdith_et_al_1_Ga_reconstruction_v1.1.zip"], "Matthews2016" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Matthews_etal_2016_Global_Plate_Model_GPC.zip"], "Merdith2017" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Merdith_etal_2017_GR.zip"], "Li2008" : [None], "Pehrsson2015" : ["https://www.geolsoc.org.uk/~/media/Files/GSL/shared/Sup_pubs/2015/18822_7.zip"], "TorsvikCocks2017" : ["http://www.earthdynamics.org/earthhistory/bookdata/CEED6.zip"], "Young2019" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Young_etal_2018_GeoscienceFrontiers/Young_etal_2018_GeoscienceFrontiers_GPlatesPlateMotionModel.zip"], "Scotese2008" : ["https://static.cambridge.org/content/id/urn:cambridge.org:id:article:S0016756818000110/resource/name/S0016756818000110sup001.zip"], "Golonka2007" : ["https://static.cambridge.org/content/id/urn:cambridge.org:id:article:S0016756818000110/resource/name/S0016756818000110sup001.zip"], "Clennett2020_M2019" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Clennett_etal_2020_G3/Clennett_etal_2020_M2019.zip"], "Clennett2020_S2013" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Clennett_etal_2020_G3/Clennett_etal_2020_S2013.zip"], "Muller2008" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Muller2008/Global_Model_Rigid_Internal_Release_2010.zip"], "Scotese2016" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Scotese2016/PALEOMAP_GlobalPlateModel.zip"], "Shephard2013" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Shephard_etal_2013_ESR.zip"], "Muller2022" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Muller_etal_2022_SE/Muller_etal_2022_SE_1Ga_Opt_PlateMotionModel_v1.1.zip"], "Cao2023" :["https://www.earthbyte.org/webdav/ftp/Data_Collections/Cao_etal_2023/1.8Ga_model_submit.zip"], "Cao2023_Opt" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Cao_etal_2023_Opt/Cao1800Opt.zip"], } return database def coastline_strings_to_include(self): strings = [ "coastline", "CEED6_LAND.gpml", # for TorsvikCocks2017 "PALEOMAP_PoliticalBoundaries", # For Scotese 2016 "coast", # for Cao2023 ] return strings def coastline_strings_to_ignore(self): strings = [ "DO_NOT", "OLD", "__MACOSX", "Clennett_2020_Coastlines", # Clennett et al. 2020 "COB_polygons_and_coastlines_combined_1000_0_Merdith_etal", # Muller et al. 2022 ] return strings def continent_strings_to_include(self): strings = [ "continent", "COBfile_1000_0_Toy_introversion", "continental", "Scotese_2008_PresentDay_ContinentalPolygons.shp", # Scotese 2008 # "Terrane", ] return strings def continent_strings_to_ignore(self): strings = [ "DO_NOT", "OLD", "__MACOSX", "Continent-ocean_boundaries", "COB", ] return strings def COB_strings_to_include(self): strings = [ "cob", "ContinentOceanBoundaries", "COBLineSegments", ] return strings def COB_strings_to_ignore(self): strings = [ "DO_NOT", "OLD", "__MACOSX", ] return stringsMethods
def COB_strings_to_ignore(self)-
Expand source code
def COB_strings_to_ignore(self): strings = [ "DO_NOT", "OLD", "__MACOSX", ] return strings def COB_strings_to_include(self)-
Expand source code
def COB_strings_to_include(self): strings = [ "cob", "ContinentOceanBoundaries", "COBLineSegments", ] return strings def coastline_strings_to_ignore(self)-
Expand source code
def coastline_strings_to_ignore(self): strings = [ "DO_NOT", "OLD", "__MACOSX", "Clennett_2020_Coastlines", # Clennett et al. 2020 "COB_polygons_and_coastlines_combined_1000_0_Merdith_etal", # Muller et al. 2022 ] return strings def coastline_strings_to_include(self)-
Expand source code
def coastline_strings_to_include(self): strings = [ "coastline", "CEED6_LAND.gpml", # for TorsvikCocks2017 "PALEOMAP_PoliticalBoundaries", # For Scotese 2016 "coast", # for Cao2023 ] return strings def continent_strings_to_ignore(self)-
Expand source code
def continent_strings_to_ignore(self): strings = [ "DO_NOT", "OLD", "__MACOSX", "Continent-ocean_boundaries", "COB", ] return strings def continent_strings_to_include(self)-
Expand source code
def continent_strings_to_include(self): strings = [ "continent", "COBfile_1000_0_Toy_introversion", "continental", "Scotese_2008_PresentDay_ContinentalPolygons.shp", # Scotese 2008 # "Terrane", ] return strings def dynamic_polygon_strings_to_ignore(self)-
Expand source code
def dynamic_polygon_strings_to_ignore(self): strings = [ "OLD", "__MACOSX", "DO_NOT", "9_Point", # Muller et al 2019 "9_Point_Density", # Clennett et al 2020 "Density", # Clennett et al 2020 "Inactive_Meshes_and_Topologies", # Clennett et al 2020 "ContinentOceanBoundaries", # Seton 2012 "Seton_etal_ESR2012_Coastline_2012", # Seton 2012 "PALEOMAP_PoliticalBoundaries", # Scotese 2016 "SimplifiedFiles", # Muller et al. 2019 (updated) "1000-410_poles", # Merdith ] return strings def dynamic_polygon_strings_to_include(self)-
Expand source code
def dynamic_polygon_strings_to_include(self): strings = [ "plate_boundaries", "PlateBoundaries", "Transform", "Divergence", "Convergence", "Topologies", "Topology", "_PP_", # for Seton 2012 #"ContinentOceanBoundaries", #"Seton_etal_ESR2012_Coastline_2012", "Deforming_Mesh", "Deforming", "Flat_Slabs", "Feature_Geometries", "boundaries", "Clennett_etal_2020_Plates", # For Clennett 2020 (M2019) "Clennett_2020_Plates", # For topologies in Clennett et al 2020 (Pacific) "Clennett_2020_Terranes", # For topologies in Clennett et al 2020 (Pacific) "Angayucham", "Farallon", "Guerrero", "Insular", "Intermontane", "Kula", "North_America", "South_America", "Western_Jurassic", "Clennett_2020_Isochrons", "Clennett_2020_Coastlines", "Clennett_2020_NAm_boundaries", "Shephard_etal_ESR2013_Global_EarthByte_2013", # For Shephard et al. 2013 "1800-1000Ma-plate-boundary_new_valid_time_and_subduction_polarity.gpml", # for Cao2023 ] return strings def netcdf4_age_grids(self, time)-
Expand source code
def netcdf4_age_grids(self, time): age_grid_links = { "Muller2019" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Muller_etal_2019_Tectonics/Muller_etal_2019_Agegrids/Muller_etal_2019_Tectonics_v2.0_netCDF/Muller_etal_2019_Tectonics_v2.0_AgeGrid-{}.nc"], "Muller2016" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Muller_etal_2016_AREPS/Muller_etal_2016_AREPS_Agegrids/Muller_etal_2016_AREPS_Agegrids_v1.17/Muller_etal_2016_AREPS_v1.17_netCDF/Muller_etal_2016_AREPS_v1.17_AgeGrid-{}.nc"], "Seton2012" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Seton_etal_2012_ESR/Seton_etal_2012_ESR_Agegrids/netCDF_0-200Ma/agegrid_{}.nc"], "Clennett2020" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Clennett_etal_2020_G3/Clennet_AgeGrids_0.1d_masked/seafloor_age_mask_{}.0Ma.nc"] } links_to_download = _find_needed_collection( self.file_collection, age_grid_links, time) return links_to_download def netcdf4_spreading_rate_grids(self, time)-
Expand source code
def netcdf4_spreading_rate_grids(self, time): spread_grid_links = { "Clennett2020" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Clennett_etal_2020_G3/Clennett_etal_2020_SpreadRate_Grids/rategrid_final_mask_{}.nc"] } links_to_download = _find_needed_collection( self.file_collection, spread_grid_links, time) return links_to_download def plate_model_valid_reconstruction_times(self)-
Expand source code
def plate_model_valid_reconstruction_times(self): database = { "Cao2020" : [0, 1000], "Muller2019" : [0, 250], "Muller2016" : [0, 230], "Clennett2020" : [0, 170], "Seton2012" : [0, 200], "Merdith2021" : [0, 1000], "Matthews2016" : [0, 410], "Merdith2017" : [0, 410], "Li2008" : [0, 410], # "Pehrsson2015" : [25], (First implement continuous rotation) "TorsvikCocks2017" : [0, 410], "Young2019" : [0, 410], "Scotese2008" : [0, 410], "Golonka2007" : [0, 410], "Clennett2020_M2019" : [0, 170], "Clennett2020_S2013" : [0, 170], "Scotese2016" : [0,410], "Shephard2013" : [0,200], "Muller2008" : [0,141], #GPlates static polygons reconstruct to this time "Muller2022" : [0,1000], "Cao2023" : [0,1800], "Cao2023_Opt" : [0,1800], } return database def plate_reconstruction_files(self)-
Expand source code
def plate_reconstruction_files(self): database = { "Cao2020" : ["https://zenodo.org/record/3854549/files/1000Myr_synthetic_tectonic_reconstructions.zip"], "Muller2019" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Muller_etal_2019_Tectonics/Muller_etal_2019_PlateMotionModel/Muller_etal_2019_PlateMotionModel_v2.0_Tectonics_Updated.zip"], "Muller2016" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Muller_etal_2016_AREPS/Muller_etal_2016_AREPS_Supplement/Muller_etal_2016_AREPS_Supplement_v1.17.zip"], "Clennett2020" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Clennett_etal_2020_G3/Global_Model_WD_Internal_Release_2019_v2_Clennett_NE_Pacific.zip"], "Seton2012" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Seton_etal_2012_ESR.zip"], #"Merdith2021" : ["https://zenodo.org/record/4485738/files/SM2_4485738_V2.zip"], "Merdith2021" : ["https://earthbyte.org/webdav/ftp/Data_Collections/Merdith_etal_2021_ESR/SM2-Merdith_et_al_1_Ga_reconstruction_v1.1.zip"], "Matthews2016" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Matthews_etal_2016_Global_Plate_Model_GPC.zip"], "Merdith2017" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Merdith_etal_2017_GR.zip"], "Li2008" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Li_etal_2008_RodiniaModel.zip"], "Pehrsson2015" : ["https://www.geolsoc.org.uk/~/media/Files/GSL/shared/Sup_pubs/2015/18822_7.zip"], "TorsvikCocks2017" : ["http://www.earthdynamics.org/earthhistory/bookdata/CEED6.zip"], "Young2019" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Young_etal_2018_GeoscienceFrontiers/Young_etal_2018_GeoscienceFrontiers_GPlatesPlateMotionModel.zip"], "Scotese2008" : ["https://static.cambridge.org/content/id/urn:cambridge.org:id:article:S0016756818000110/resource/name/S0016756818000110sup001.zip"], "Golonka2007" : ["https://static.cambridge.org/content/id/urn:cambridge.org:id:article:S0016756818000110/resource/name/S0016756818000110sup001.zip"], "Clennett2020_M2019" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Clennett_etal_2020_G3/Clennett_etal_2020_M2019.zip"], "Clennett2020_S2013" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Clennett_etal_2020_G3/Clennett_etal_2020_S2013.zip"], "Muller2008" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Muller2008/Global_Model_Rigid_Internal_Release_2010.zip"], "Scotese2016" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Scotese2016/PALEOMAP_GlobalPlateModel.zip"], "Shephard2013" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Shephard_etal_2013_ESR.zip"], "Muller2022" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Muller_etal_2022_SE/Muller_etal_2022_SE_1Ga_Opt_PlateMotionModel_v1.1.zip"], "Cao2023" :["https://www.earthbyte.org/webdav/ftp/Data_Collections/Cao_etal_2023/1.8Ga_model_submit.zip"], "Cao2023_Opt" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Cao_etal_2023_Opt/Cao1800Opt.zip"], } return database def rotation_strings_to_ignore(self)-
Expand source code
def rotation_strings_to_ignore(self): strings = [ "OLD", "__MACOSX", "DO_NOT", "Blocks_crossing_Poles" ] return strings def rotation_strings_to_include(self)-
Expand source code
def rotation_strings_to_include(self): strings = [ "Muller2022 1000_0_rotfile_Merdith_et_al_optimised.rot", # For Muller et al. 2022 ] return strings def static_polygon_strings_to_ignore(self)-
Expand source code
def static_polygon_strings_to_ignore(self): strings = [ "DO_NOT", "OLD", "__MACOSX", "Global_Model_WD_Internal_Release_2019_v2_Clennett_NE_Pacific/StaticGeometries/StaticPolygons/Global_EarthByte_GPlates_PresentDay_StaticPlatePolygons.shp" # Clennett 2020 ] return strings def static_polygon_strings_to_include(self)-
Expand source code
def static_polygon_strings_to_include(self): strings = [ "StaticPolygon", "StaticPolygons", "Static_Polygon", "StaticPlatePolygons_", "RodiniaBlocks_WithPlateIDColumnAndIDs", # "PlatePolygons.shp", "CEED6_TERRANES.shp", "CEED6_MICROCONTINENTS.shp", "CEED6_LAND.gpml", "Scotese_2008_PresentDay_ContinentalPolygons", # Scotese 2008 "Golonka_2007_PresentDay_ContinentalPolygons.shp", # Golonka 2007 "PALEOMAP_PlatePolygons.gpml", # For Scotese 2016 ] return strings def topology_geometries(self)-
Expand source code
def topology_geometries(self): database = { "Cao2020" : ["https://zenodo.org/record/3854549/files/1000Myr_synthetic_tectonic_reconstructions.zip"], "Muller2019" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Muller_etal_2019_Tectonics/Muller_etal_2019_PlateMotionModel/Muller_etal_2019_PlateMotionModel_v2.0_Tectonics_Updated.zip"], "Muller2016" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Muller_etal_2016_AREPS/Muller_etal_2016_AREPS_Supplement/Muller_etal_2016_AREPS_Supplement_v1.17.zip"], "Clennett2020" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Clennett_etal_2020_G3/Global_Model_WD_Internal_Release_2019_v2_Clennett_NE_Pacific.zip"], "Seton2012" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Seton_etal_2012_ESR.zip"], #"Merdith2021" : ["https://zenodo.org/record/4485738/files/SM2_4485738_V2.zip"], "Merdith2021" : ["https://earthbyte.org/webdav/ftp/Data_Collections/Merdith_etal_2021_ESR/SM2-Merdith_et_al_1_Ga_reconstruction_v1.1.zip"], "Matthews2016" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Matthews_etal_2016_Global_Plate_Model_GPC.zip"], "Merdith2017" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Merdith_etal_2017_GR.zip"], "Li2008" : [None], "Pehrsson2015" : ["https://www.geolsoc.org.uk/~/media/Files/GSL/shared/Sup_pubs/2015/18822_7.zip"], "TorsvikCocks2017" : ["http://www.earthdynamics.org/earthhistory/bookdata/CEED6.zip"], "Young2019" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Young_etal_2018_GeoscienceFrontiers/Young_etal_2018_GeoscienceFrontiers_GPlatesPlateMotionModel.zip"], "Scotese2008" : ["https://static.cambridge.org/content/id/urn:cambridge.org:id:article:S0016756818000110/resource/name/S0016756818000110sup001.zip"], "Golonka2007" : ["https://static.cambridge.org/content/id/urn:cambridge.org:id:article:S0016756818000110/resource/name/S0016756818000110sup001.zip"], "Clennett2020_M2019" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Clennett_etal_2020_G3/Clennett_etal_2020_M2019.zip"], "Clennett2020_S2013" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Clennett_etal_2020_G3/Clennett_etal_2020_S2013.zip"], "Muller2008" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Muller2008/Global_Model_Rigid_Internal_Release_2010.zip"], "Scotese2016" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Scotese2016/PALEOMAP_GlobalPlateModel.zip"], "Shephard2013" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Shephard_etal_2013_ESR.zip"], "Muller2022" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Muller_etal_2022_SE/Muller_etal_2022_SE_1Ga_Opt_PlateMotionModel_v1.1.zip"], "Cao2023" :["https://www.earthbyte.org/webdav/ftp/Data_Collections/Cao_etal_2023/1.8Ga_model_submit.zip"], "Cao2023_Opt" : ["https://www.earthbyte.org/webdav/ftp/Data_Collections/Cao_etal_2023_Opt/Cao1800Opt.zip"], } return database
class DataServer (file_collection, verbose=True)-
Uses Pooch to download plate reconstruction feature data from plate models and other studies that are stored on web servers (e.g. EarthByte's webDAV server).
If the
DataServerobject and its methods are called for the first time, i.e. by:# string identifier to access the Muller et al. 2019 model gDownload = gplately.download.DataServer("Muller2019")all requested files are downloaded into the user's 'gplately' cache folder only once. If the same object and method(s) are re-run, the files will be re-accessed from the cache provided they have not been moved or deleted.
Currently,
DataServersupports a number of plate reconstruction models.
Model name string Identifier Rot. files Topology features Static polygons Coast-lines Cont-inents COB- Age grids SR grids Muller2019 ✅ ✅ ✅ ✅ ✅ ✅ ✅ ❌ Muller2016 ✅ ✅ ✅ ✅ ❌ ❌ ✅ ❌ Merdith2021 ✅ ✅ ✅ ✅ ✅ ❌ ❌ ❌ Cao2020 ✅ ✅ ✅ ✅ ✅ ❌ ❌ ❌ Clennett2020 ✅ ✅ ✅ ✅ ✅ ❌ ✅ ✅ Seton2012 ✅ ✅ ❌ ✅ ❌ ✅ ✅ ❌ Matthews2016 ✅ ✅ ✅ ✅ ✅ ❌ ❌ ❌ Merdith2017 ✅ ✅ ❌ ❌ ❌ ❌ ❌ ❌ Li2008 ✅ ✅ ❌ ❌ ❌ ❌ ❌ ❌ Pehrsson2015 ✅ ✅ ❌ ❌ ❌ ❌ ❌ ❌ TorsvikCocks2017 ✅ ❌ ❌ ✅ ❌ ❌ ❌ ❌ Young2019 ✅ ✅ ✅ ✅ ✅ ❌ ❌ ❌ Scotese2008 ✅ ✅ ❌ ❌ ✅ ❌ ❌ ❌ Clennett2020_M19 ✅ ✅ ❌ ✅ ✅ ❌ ❌ ❌ Clennett2020_S13 ✅ ✅ ❌ ✅ ✅ ❌ ❌ ❌ Muller2008 ✅ ❌ ✅ ❌ ❌ ❌ ❌ ❌ Muller2022 ✅ ✅ ✅ ✅ ✅ ✅ ❌ ❌ Scotese2016 ✅ ❌ ✅ ✅ ❌ ❌ ❌ ❌ Shephard2013 ✅ ✅ ✅ ✅ ❌ ❌ ❌ ❌
To call the object, supply a model name string Identifier,
file_collection, from one of the following models:-
file_collection =
Muller2019Information
- Downloadable files: a
rotation_model,topology_features,static_polygons,coastlines,continents,COBs, and seafloorage_gridsfrom 0 to 250 Ma. - Maximum reconstruction time: 250 Ma
Citations
Müller, R. D., Zahirovic, S., Williams, S. E., Cannon, J., Seton, M., Bower, D. J., Tetley, M. G., Heine, C., Le Breton, E., Liu, S., Russell, S. H. J., Yang, T., Leonard, J., and Gurnis, M. (2019), A global plate model including lithospheric deformation along major rifts and orogens since the Triassic. Tectonics, vol. 38, https://doi.org/10.1029/2018TC005462.
- Downloadable files: a
-
Müller et al. 2016:
file_collection =
Muller2016Information
- Downloadable files: a
rotation_model,topology_features,static_polygons,coastlines, and seafloorage_gridsfrom 0-230 Ma. - Maximum reconstruction time: 230 Ma
Citations
- Müller R.D., Seton, M., Zahirovic, S., Williams, S.E., Matthews, K.J., Wright, N.M., Shephard, G.E., Maloney, K.T., Barnett-Moore, N., Hosseinpour, M., Bower, D.J., Cannon, J., InPress. Ocean basin evolution and global-scale plate reorganization events since Pangea breakup, Annual Review of Earth and Planetary Sciences, Vol 44, 107-138. DOI: 10.1146/annurev-earth-060115-012211.
- Downloadable files: a
-
file_collection =
Merdith2021Information
- Downloadable files: a
rotation_model,topology_features,static_polygons,coastlinesandcontinents. - Maximum reconstruction time: 1 Ga (however,
PlotTopologiescorrectly visualises up to 410 Ma)
Citations:
- Merdith et al. (in review), 'A continuous, kinematic full-plate motion model from 1 Ga to present'.
- Andrew Merdith. (2020). Plate model for 'Extending Full-Plate Tectonic Models into Deep Time: Linking the Neoproterozoic and the Phanerozoic ' (1.1b) [Data set]. Zenodo. https://doi.org/10.5281/zenodo.4485738
- Downloadable files: a
-
Cao et al. 2020:
file_collection =
Cao2020Information
- Downloadable files:
rotation_model,topology_features,static_polygons,coastlinesandcontinents. - Maximum reconstruction time: 1 Ga
Citations
- Toy Billion-year reconstructions from Cao et al (2020). Coupled Evolution of Plate Tectonics and Basal Mantle Structure Tectonics, doi: 10.1029/2020GC009244
- Downloadable files:
-
Clennett et al. 2020 :
file_collection =
Clennett2020Information
- Downloadable files:
rotation_model,topology_features,static_polygons,coastlinesandcontinents - Maximum reconstruction time: 170 Ma
Citations
- Mather, B., Müller, R.D.,; Alfonso, C.P., Seton, M., 2021, Kimberlite eruption driven by slab flux and subduction angle. DOI: 10.5281/zenodo.5769002
- Downloadable files:
-
Seton et al. 2012:
file_collection =
Seton2012Information
- Downloadable files:
rotation_model,topology_features,coastlines,COBs, and paleo-age grids (0-200 Ma) - Maximum reconstruction time: 200 Ma
Citations
- M. Seton, R.D. Müller, S. Zahirovic, C. Gaina, T.H. Torsvik, G. Shephard, A. Talsma, M. Gurnis, M. Turner, S. Maus, M. Chandler, Global continental and ocean basin reconstructions since 200 Ma, Earth-Science Reviews, Volume 113, Issues 3-4, July 2012, Pages 212-270, ISSN 0012-8252, 10.1016/j.earscirev.2012.03.002.
- Downloadable files:
-
Matthews et al. 2016:
file_collection =
Matthews2016Information
- Downloadable files:
rotation_model,topology_features,static_polygons,coastlines, andcontinents - Maximum reconstruction time(s): 410-250 Ma, 250-0 Ma
Citations
- Matthews, K.J., Maloney, K.T., Zahirovic, S., Williams, S.E., Seton, M., and Müller, R.D. (2016). Global plate boundary evolution and kinematics since the late Paleozoic, Global and Planetary Change, 146, 226-250. DOI: 10.1016/j.gloplacha.2016.10.002
- Downloadable files:
-
Merdith et al. 2017:
file_collection =
Merdith2017Information
- Downloadable files:
rotation_filesandtopology_features - Maximum reconstruction time: 410 Ma
Citations
- Merdith, A., Collins, A., Williams, S., Pisarevskiy, S., Foden, J., Archibald, D. and Blades, M. et al. 2016. A full-plate global reconstruction of the Neoproterozoic. Gondwana Research. 50: pp. 84-134. DOI: 10.1016/j.gr.2017.04.001
- Downloadable files:
-
Li et al. 2008:
file_collection =
Li2008Information
- Downloadable files:
rotation_modelandstatic_polygons - Maximum reconstruction time: 410 Ma
Citations
- Rodinia reconstruction from Li et al (2008), Assembly, configuration, and break-up history of Rodinia: A synthesis. Precambrian Research. 160. 179-210. DOI: 10.1016/j.precamres.2007.04.021.
- Downloadable files:
-
Pehrsson et al. 2015:
file_collection =
Pehrsson2015Information
- Downloadable files:
rotation_modelandstatic_polygons - Maximum reconstruction time: N/A
Citations
- Pehrsson, S.J., Eglington, B.M., Evans, D.A.D., Huston, D., and Reddy, S.M., (2015), Metallogeny and its link to orogenic style during the Nuna supercontinent cycle. Geological Society, London, Special Publications, 424, 83-94. DOI: https://doi.org/10.1144/SP424.5
- Downloadable files:
-
Torsvik and Cocks et al. 2017:
file_collection =
TorsvikCocks2017Information
- Downloadable files:
rotation_model, andcoastlines - Maximum reconstruction time: 410 Ma
Citations
- Torsvik, T., & Cocks, L. (2016). Earth History and Palaeogeography. Cambridge: Cambridge University Press. doi:10.1017/9781316225523
- Downloadable files:
-
Young et al. 2019:
file_collection =
Young2019Information
- Downloadable files:
rotation_model,topology_features,static_polygons,coastlinesandcontinents. - Maximum reconstruction time: 410-250 Ma, 250-0 Ma
Citations
- Young, A., Flament, N., Maloney, K., Williams, S., Matthews, K., Zahirovic, S., Müller, R.D., (2019), Global kinematics of tectonic plates and subduction zones since the late Paleozoic Era, Geoscience Frontiers, Volume 10, Issue 3, pp. 989-1013, ISSN 1674-9871, DOI: https://doi.org/10.1016/j.gsf.2018.05.011.
- Downloadable files:
-
Scotese et al. 2008:
file_collection =
Scotese2008Information
- Downloadable files:
rotation_model,static_polygons, andcontinents - Maximum reconstruction time:
Citations
- Scotese, C.R. 2008. The PALEOMAP Project PaleoAtlas for ArcGIS, Volume 2, Cretaceous paleogeographic and plate tectonic reconstructions. PALEOMAP Project, Arlington, Texas.
- Downloadable files:
-
Golonka et al. 2007:
file_collection =
Golonka2007Information
- Downloadable files:
rotation_model,static_polygons, andcontinents - Maximum reconstruction time: 410 Ma
Citations
- Golonka, J. 2007. Late Triassic and Early Jurassic palaeogeography of the world. Palaeogeography, Palaeoclimatology, Palaeoecology 244(1–4), 297–307.
- Downloadable files:
-
Clennett et al. 2020 (based on Müller et al. 2019):
file_collection =
Clennett2020_M2019Information
- Downloadable files:
rotation_model,topology_features,continentsandcoastlines - Maximum reconstruction time: 170 Ma
Citations
- Clennett, E.J., Sigloch, K., Mihalynuk, M.G., Seton, M., Henderson, M.A., Hosseini, K., Mohammadzaheri, A., Johnston, S.T., Müller, R.D., (2020), A Quantitative Tomotectonic Plate Reconstruction of Western North America and the Eastern Pacific Basin. Geochemistry, Geophysics, Geosystems, 21, e2020GC009117. DOI: https://doi.org/10.1029/2020GC009117
- Downloadable files:
-
Clennett et al. 2020 (rigid topological model based on Shephard et al, 2013):
file_collection =
Clennett2020_S2013Information
- Downloadable files:
rotation_model,topology_features,continentsandcoastlines - Maximum reconstruction time: 170
Citations
- Clennett, E.J., Sigloch, K., Mihalynuk, M.G., Seton, M., Henderson, M.A., Hosseini, K., Mohammadzaheri, A., Johnston, S.T., Müller, R.D., (2020), A Quantitative Tomotectonic Plate Reconstruction of Western North America and the Eastern Pacific Basin. Geochemistry, Geophysics, Geosystems, 21, e2020GC009117. DOI: https://doi.org/10.1029/2020GC009117
- Downloadable files:
-
Müller et al. 2008:
file_collection =
Muller2008Information
- Downloadable files:
rotation_model,static_polygons - Maximum reconstruction time: 141 Ma
Citations
- Müller, R. D., Sdrolias, M., Gaina, C., & Roest, W. R. (2008). Age, spreading rates, and spreading asymmetry of the world's ocean crust. Geochemistry, Geophysics, Geosystems, 9(4).
- Downloadable files:
-
Müller et al. 2022:
file_collection =
Muller2022Information
- Downloadable files:
rotation_model,topology_features,static_polygons,continents,coastlinesandCOBs - Maximum reconstruction time: 1000 Ma
Citations
- Müller, R. D., Flament, N., Cannon, J., Tetley, M. G., Williams, S. E., Cao, X., Bodur, Ö. F., Zahirovic, S., and Merdith, A.: A tectonic-rules-based mantle reference frame since 1 billion years ago – implications for supercontinent cycles and plate–mantle system evolution, Solid Earth, 13, 1127–1159, https://doi.org/10.5194/se-13-1127-2022, 2022.
- Downloadable files:
-
Scotese 2016:
file_collection =
Scotese2016Information
- Downloadable files:
rotation_model,static_polygons,coastlines - Maximum reconstruction time: 410 Ma
Citations
- Scotese, C.R., 2016. PALEOMAP PaleoAtlas for GPlates and the PaleoData Plotter Program, PALEOMAP Project, http://www.earthbyte.org/paleomappaleoatlas-for-gplates/
- Downloadable files:
-
Shephard et al. 2013:
file_collection =
Shephard2013Information
- Downloadable files:
rotation_model,topology_features,static_polygons,coastlines - Maximum reconstruction time: 200 Ma
Citations
-
Shephard, G.E., Müller, R.D., and Seton, M., 2013. The tectonic evolution of the Arctic since Pangea breakup: Integrating constraints from surface geology and geophysics with mantle structure. Earth-Science Reviews, Volume 124 p.148-183. doi:10.1016/j.earscirev.2013.05.012 (http://www.sciencedirect.com/science/article/pii/S0012825213001104)
-
M. Seton, R.D. Müller, S. Zahirovic, C. Gaina, T.H. Torsvik, G. Shephard, A. Talsma, M. Gurnis, M. Turner, S. Maus, M. Chandler, Global continental and ocean basin reconstructions since 200 Ma, Earth-Science Reviews, Volume 113, p.212-270 doi:10.1016/j.earscirev.2012.03.002. (http://www.sciencedirect.com/science/article/pii/S0012825212000311)
Parameters
file_collection : str name of file collection to use
verbose : bool, default True Toggle print messages regarding server/internet connection status, file availability etc.
- Downloadable files:
Expand source code
class DataServer(object): """Uses [Pooch](https://www.fatiando.org/pooch/latest/) to download plate reconstruction feature data from plate models and other studies that are stored on web servers (e.g. EarthByte's [webDAV server](https://www.earthbyte.org/webdav/ftp/Data_Collections/)). If the `DataServer` object and its methods are called for the first time, i.e. by: # string identifier to access the Muller et al. 2019 model gDownload = gplately.download.DataServer("Muller2019") all requested files are downloaded into the user's 'gplately' cache folder only _once_. If the same object and method(s) are re-run, the files will be re-accessed from the cache provided they have not been moved or deleted. Currently, `DataServer` supports a number of plate reconstruction models. ------------------ | **Model name string Identifier** | **Rot. files** | **Topology features** | **Static polygons** | **Coast-lines** | **Cont-inents** | **COB-** | **Age grids** | **SR grids** | |:--------------------------------:|:--------------:|:---------------------:|:-------------------:|:---------------:|:---------------:|:--------:|:-------------:|:------------:| | Muller2019 | ✅ | ✅ | ✅ | ✅ | ✅ | ✅ | ✅ | ❌ | | Muller2016 | ✅ | ✅ | ✅ | ✅ | ❌ | ❌ | ✅ | ❌ | | Merdith2021 | ✅ | ✅ | ✅ | ✅ | ✅ | ❌ | ❌ | ❌ | | Cao2020 | ✅ | ✅ | ✅ | ✅ | ✅ | ❌ | ❌ | ❌ | | Clennett2020 | ✅ | ✅ | ✅ | ✅ | ✅ | ❌ | ✅ | ✅ | | Seton2012 | ✅ | ✅ | ❌ | ✅ | ❌ | ✅ | ✅ | ❌ | | Matthews2016 | ✅ | ✅ | ✅ | ✅ | ✅ | ❌ | ❌ | ❌ | | Merdith2017 | ✅ | ✅ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ | | Li2008 | ✅ | ✅ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ | | Pehrsson2015 | ✅ | ✅ | ❌ | ❌ | ❌ | ❌ | ❌ | ❌ | | TorsvikCocks2017 | ✅ | ❌ | ❌ | ✅ | ❌ | ❌ | ❌ | ❌ | | Young2019 | ✅ | ✅ | ✅ | ✅ | ✅ | ❌ | ❌ | ❌ | | Scotese2008 | ✅ | ✅ | ❌ | ❌ | ✅ | ❌ | ❌ | ❌ | | Clennett2020_M19 | ✅ | ✅ | ❌ | ✅ | ✅ | ❌ | ❌ | ❌ | | Clennett2020_S13 | ✅ | ✅ | ❌ | ✅ | ✅ | ❌ | ❌ | ❌ | | Muller2008 | ✅ | ❌ | ✅ | ❌ | ❌ | ❌ | ❌ | ❌ | | Muller2022 | ✅ | ✅ | ✅ | ✅ | ✅ | ✅ | ❌ | ❌ | | Scotese2016 | ✅ | ❌ | ✅ | ✅ | ❌ | ❌ | ❌ | ❌ | | Shephard2013 | ✅ | ✅ | ✅ | ✅ | ❌ | ❌ | ❌ | ❌ | ------------------ To call the object, supply a model name string Identifier, `file_collection`, from one of the following models: * __[Müller et al. 2019](https://www.earthbyte.org/muller-et-al-2019-deforming-plate-reconstruction-and-seafloor-age-grids-tectonics/):__ file_collection = `Muller2019` Information ----------- * Downloadable files: a `rotation_model`, `topology_features`, `static_polygons`, `coastlines`, `continents`, `COBs`, and seafloor `age_grids` from 0 to 250 Ma. * Maximum reconstruction time: 250 Ma Citations --------- Müller, R. D., Zahirovic, S., Williams, S. E., Cannon, J., Seton, M., Bower, D. J., Tetley, M. G., Heine, C., Le Breton, E., Liu, S., Russell, S. H. J., Yang, T., Leonard, J., and Gurnis, M. (2019), A global plate model including lithospheric deformation along major rifts and orogens since the Triassic. Tectonics, vol. 38, https://doi.org/10.1029/2018TC005462. * __Müller et al. 2016__: file_collection = `Muller2016` Information ----------- * Downloadable files: a `rotation_model`, `topology_features`, `static_polygons`, `coastlines`, and seafloor `age_grids` from 0-230 Ma. * Maximum reconstruction time: 230 Ma Citations --------- * Müller R.D., Seton, M., Zahirovic, S., Williams, S.E., Matthews, K.J., Wright, N.M., Shephard, G.E., Maloney, K.T., Barnett-Moore, N., Hosseinpour, M., Bower, D.J., Cannon, J., InPress. Ocean basin evolution and global-scale plate reorganization events since Pangea breakup, Annual Review of Earth and Planetary Sciences, Vol 44, 107-138. DOI: 10.1146/annurev-earth-060115-012211. * __[Merdith et al. 2021](https://zenodo.org/record/4485738#.Yhrm8hNBzA0)__: file_collection = `Merdith2021` Information ----------- * Downloadable files: a `rotation_model`, `topology_features`, `static_polygons`, `coastlines` and `continents`. * Maximum reconstruction time: 1 Ga (however, `PlotTopologies` correctly visualises up to 410 Ma) Citations: ---------- * Merdith et al. (in review), 'A continuous, kinematic full-plate motion model from 1 Ga to present'. * Andrew Merdith. (2020). Plate model for 'Extending Full-Plate Tectonic Models into Deep Time: Linking the Neoproterozoic and the Phanerozoic ' (1.1b) [Data set]. Zenodo. https://doi.org/10.5281/zenodo.4485738 * __Cao et al. 2020__: file_collection = `Cao2020` Information ----------- * Downloadable files: `rotation_model`, `topology_features`, `static_polygons`, `coastlines` and `continents`. * Maximum reconstruction time: 1 Ga Citations --------- * Toy Billion-year reconstructions from Cao et al (2020). Coupled Evolution of Plate Tectonics and Basal Mantle Structure Tectonics, doi: 10.1029/2020GC009244 - __Clennett et al. 2020__ : file_collection = `Clennett2020` Information ----------- * Downloadable files: `rotation_model`, `topology_features`, `static_polygons`, `coastlines` and `continents` * Maximum reconstruction time: 170 Ma Citations --------- * Mather, B., Müller, R.D.,; Alfonso, C.P., Seton, M., 2021, Kimberlite eruption driven by slab flux and subduction angle. DOI: 10.5281/zenodo.5769002 - __Seton et al. 2012__: file_collection = `Seton2012` Information ----------- * Downloadable files: `rotation_model`, `topology_features`, `coastlines`, `COBs`, and paleo-age grids (0-200 Ma) * Maximum reconstruction time: 200 Ma Citations --------- * M. Seton, R.D. Müller, S. Zahirovic, C. Gaina, T.H. Torsvik, G. Shephard, A. Talsma, M. Gurnis, M. Turner, S. Maus, M. Chandler, Global continental and ocean basin reconstructions since 200 Ma, Earth-Science Reviews, Volume 113, Issues 3-4, July 2012, Pages 212-270, ISSN 0012-8252, 10.1016/j.earscirev.2012.03.002. - __Matthews et al. 2016__: file_collection = `Matthews2016` Information ----------- * Downloadable files: `rotation_model`, `topology_features`, `static_polygons`, `coastlines`, and `continents` * Maximum reconstruction time(s): 410-250 Ma, 250-0 Ma Citations --------- * Matthews, K.J., Maloney, K.T., Zahirovic, S., Williams, S.E., Seton, M., and Müller, R.D. (2016). Global plate boundary evolution and kinematics since the late Paleozoic, Global and Planetary Change, 146, 226-250. DOI: 10.1016/j.gloplacha.2016.10.002 - __Merdith et al. 2017__: file_collection = `Merdith2017` Information ----------- * Downloadable files: `rotation_files` and `topology_features` * Maximum reconstruction time: 410 Ma Citations --------- * Merdith, A., Collins, A., Williams, S., Pisarevskiy, S., Foden, J., Archibald, D. and Blades, M. et al. 2016. A full-plate global reconstruction of the Neoproterozoic. Gondwana Research. 50: pp. 84-134. DOI: 10.1016/j.gr.2017.04.001 - __Li et al. 2008__: file_collection = `Li2008` Information ----------- * Downloadable files: `rotation_model` and `static_polygons` * Maximum reconstruction time: 410 Ma Citations --------- * Rodinia reconstruction from Li et al (2008), Assembly, configuration, and break-up history of Rodinia: A synthesis. Precambrian Research. 160. 179-210. DOI: 10.1016/j.precamres.2007.04.021. - __Pehrsson et al. 2015__: file_collection = `Pehrsson2015` Information ----------- * Downloadable files: `rotation_model` and `static_polygons` * Maximum reconstruction time: N/A Citations --------- * Pehrsson, S.J., Eglington, B.M., Evans, D.A.D., Huston, D., and Reddy, S.M., (2015), Metallogeny and its link to orogenic style during the Nuna supercontinent cycle. Geological Society, London, Special Publications, 424, 83-94. DOI: https://doi.org/10.1144/SP424.5 - __Torsvik and Cocks et al. 2017__: file_collection = `TorsvikCocks2017` Information ----------- * Downloadable files: `rotation_model`, and `coastlines` * Maximum reconstruction time: 410 Ma Citations --------- * Torsvik, T., & Cocks, L. (2016). Earth History and Palaeogeography. Cambridge: Cambridge University Press. doi:10.1017/9781316225523 - __Young et al. 2019__: file_collection = `Young2019` Information ----------- * Downloadable files: `rotation_model`, `topology_features`, `static_polygons`, `coastlines` and `continents`. * Maximum reconstruction time: 410-250 Ma, 250-0 Ma Citations --------- * Young, A., Flament, N., Maloney, K., Williams, S., Matthews, K., Zahirovic, S., Müller, R.D., (2019), Global kinematics of tectonic plates and subduction zones since the late Paleozoic Era, Geoscience Frontiers, Volume 10, Issue 3, pp. 989-1013, ISSN 1674-9871, DOI: https://doi.org/10.1016/j.gsf.2018.05.011. - __Scotese et al. 2008__: file_collection = `Scotese2008` Information ----------- * Downloadable files: `rotation_model`, `static_polygons`, and `continents` * Maximum reconstruction time: Citations --------- * Scotese, C.R. 2008. The PALEOMAP Project PaleoAtlas for ArcGIS, Volume 2, Cretaceous paleogeographic and plate tectonic reconstructions. PALEOMAP Project, Arlington, Texas. - __Golonka et al. 2007__: file_collection = `Golonka2007` Information ----------- * Downloadable files: `rotation_model`, `static_polygons`, and `continents` * Maximum reconstruction time: 410 Ma Citations --------- * Golonka, J. 2007. Late Triassic and Early Jurassic palaeogeography of the world. Palaeogeography, Palaeoclimatology, Palaeoecology 244(1–4), 297–307. - __Clennett et al. 2020 (based on Müller et al. 2019)__: file_collection = `Clennett2020_M2019` Information ----------- * Downloadable files: `rotation_model`, `topology_features`, `continents` and `coastlines` * Maximum reconstruction time: 170 Ma Citations --------- * Clennett, E.J., Sigloch, K., Mihalynuk, M.G., Seton, M., Henderson, M.A., Hosseini, K., Mohammadzaheri, A., Johnston, S.T., Müller, R.D., (2020), A Quantitative Tomotectonic Plate Reconstruction of Western North America and the Eastern Pacific Basin. Geochemistry, Geophysics, Geosystems, 21, e2020GC009117. DOI: https://doi.org/10.1029/2020GC009117 - __Clennett et al. 2020 (rigid topological model based on Shephard et al, 2013)__: file_collection = `Clennett2020_S2013` Information ----------- * Downloadable files: `rotation_model`, `topology_features`, `continents` and `coastlines` * Maximum reconstruction time: 170 Citations --------- * Clennett, E.J., Sigloch, K., Mihalynuk, M.G., Seton, M., Henderson, M.A., Hosseini, K., Mohammadzaheri, A., Johnston, S.T., Müller, R.D., (2020), A Quantitative Tomotectonic Plate Reconstruction of Western North America and the Eastern Pacific Basin. Geochemistry, Geophysics, Geosystems, 21, e2020GC009117. DOI: https://doi.org/10.1029/2020GC009117 - __Müller et al. 2008__: file_collection = `Muller2008` Information ----------- * Downloadable files: `rotation_model`, `static_polygons` * Maximum reconstruction time: 141 Ma Citations --------- * Müller, R. D., Sdrolias, M., Gaina, C., & Roest, W. R. (2008). Age, spreading rates, and spreading asymmetry of the world's ocean crust. Geochemistry, Geophysics, Geosystems, 9(4). - __Müller et al. 2022__: file_collection = `Muller2022` Information ----------- * Downloadable files: `rotation_model`, `topology_features`, `static_polygons`, `continents`, `coastlines` and `COBs` * Maximum reconstruction time: 1000 Ma Citations --------- * Müller, R. D., Flament, N., Cannon, J., Tetley, M. G., Williams, S. E., Cao, X., Bodur, Ö. F., Zahirovic, S., and Merdith, A.: A tectonic-rules-based mantle reference frame since 1 billion years ago – implications for supercontinent cycles and plate–mantle system evolution, Solid Earth, 13, 1127–1159, https://doi.org/10.5194/se-13-1127-2022, 2022. - __Scotese 2016__: file_collection = `Scotese2016` Information ----------- * Downloadable files: `rotation_model`, `static_polygons`, `coastlines` * Maximum reconstruction time: 410 Ma Citations --------- * Scotese, C.R., 2016. PALEOMAP PaleoAtlas for GPlates and the PaleoData Plotter Program, PALEOMAP Project, http://www.earthbyte.org/paleomappaleoatlas-for-gplates/ - __Shephard et al. 2013__: file_collection = `Shephard2013` Information ----------- * Downloadable files: `rotation_model`, `topology_features`, `static_polygons`, `coastlines` * Maximum reconstruction time: 200 Ma Citations --------- * Shephard, G.E., Müller, R.D., and Seton, M., 2013. The tectonic evolution of the Arctic since Pangea breakup: Integrating constraints from surface geology and geophysics with mantle structure. Earth-Science Reviews, Volume 124 p.148-183. doi:10.1016/j.earscirev.2013.05.012 (http://www.sciencedirect.com/science/article/pii/S0012825213001104) * M. Seton, R.D. Müller, S. Zahirovic, C. Gaina, T.H. Torsvik, G. Shephard, A. Talsma, M. Gurnis, M. Turner, S. Maus, M. Chandler, Global continental and ocean basin reconstructions since 200 Ma, Earth-Science Reviews, Volume 113, p.212-270 doi:10.1016/j.earscirev.2012.03.002. (http://www.sciencedirect.com/science/article/pii/S0012825212000311) Parameters ---------- file_collection : str name of file collection to use verbose : bool, default True Toggle print messages regarding server/internet connection status, file availability etc. """ def __init__(self, file_collection, verbose=True): self.file_collection = file_collection.capitalize() self.data_collection = DataCollection(self.file_collection) if str(type(verbose)) == "<class 'bool'>": self.verbose = verbose else: raise ValueError("The verbose toggle must be of Boolean type, not {}".format(type(verbose))) def get_plate_reconstruction_files(self): """Downloads and constructs a `rotation model`, a set of `topology_features` and and a set of `static_polygons` needed to call the `PlateReconstruction` object. Returns ------- rotation_model : instance of <pygplates.RotationModel> A rotation model to query equivalent and/or relative topological plate rotations from a time in the past relative to another time in the past or to present day. topology_features : instance of <pygplates.FeatureCollection> Point, polyline and/or polygon feature data that are reconstructable through geological time. static_polygons : instance of <pygplates.FeatureCollection> Present-day polygons whose shapes do not change through geological time. They are used to cookie-cut dynamic polygons into identifiable topological plates (assigned an ID) according to their present-day locations. Notes ----- This method accesses the plate reconstruction model ascribed to the `file_collection` string passed into the `DataServer` object. For example, if the object was called with `"Muller2019"`: gDownload = gplately.download.DataServer("Muller2019") rotation_model, topology_features, static_polygons = gDownload.get_plate_reconstruction_files() the method will download a `rotation_model`, `topology_features` and `static_polygons` from the Müller et al. (2019) plate reconstruction model. Once the reconstruction objects are returned, they can be passed into: model = gplately.reconstruction.PlateReconstruction(rotation_model, topology_features, static_polygons) * Note: If the requested plate model does not have a certain file(s), a message will be printed to alert the user. For example, using `get_plate_reconstruction_files()` for the Torsvik and Cocks (2017) plate reconstruction model yields the printed message: No topology features in TorsvikCocks2017. No FeatureCollection created - unable to plot trenches, ridges and transforms. No continent-ocean boundaries in TorsvikCocks2017. """ verbose = self.verbose rotation_filenames = [] rotation_model = [] topology_filenames = [] topology_features = _FeatureCollection() static_polygons= _FeatureCollection() static_polygon_filenames = [] # Locate all plate reconstruction files from GPlately's DataCollection database = DataCollection.plate_reconstruction_files(self) # Set to true if we find the given collection in our database found_collection = False for collection, url in database.items(): # Only continue if the user's chosen collection exists in our database if self.file_collection.lower() == collection.lower(): found_collection = True if len(url) == 1: fnames = _collection_sorter( download_from_web(url[0], verbose, model_name=self.file_collection), self.file_collection ) rotation_filenames = _collect_file_extension( _str_in_folder( _str_in_filename(fnames, strings_to_include=DataCollection.rotation_strings_to_include(self), strings_to_ignore=DataCollection.rotation_strings_to_ignore(self), file_collection=self.file_collection, file_collection_sensitive=True ), strings_to_ignore=DataCollection.rotation_strings_to_ignore(self) ), [".rot"] ) #print(rotation_filenames) rotation_model = _RotationModel(rotation_filenames) topology_filenames = _collect_file_extension( _str_in_folder( _str_in_filename(fnames, strings_to_include=DataCollection.dynamic_polygon_strings_to_include(self), strings_to_ignore=DataCollection.dynamic_polygon_strings_to_ignore(self), file_collection=self.file_collection, file_collection_sensitive=False, ), strings_to_ignore=DataCollection.dynamic_polygon_strings_to_ignore(self) ), [".gpml", ".gpmlz"] ) #print(topology_filenames) for file in topology_filenames: topology_features.add(_FeatureCollection(file)) static_polygon_filenames = _check_gpml_or_shp( _str_in_folder( _str_in_filename(fnames, strings_to_include=DataCollection.static_polygon_strings_to_include(self), strings_to_ignore=DataCollection.static_polygon_strings_to_ignore(self), file_collection=self.file_collection, file_collection_sensitive=False ), strings_to_ignore=DataCollection.static_polygon_strings_to_ignore(self) ) ) #print(static_polygon_filenames) for stat in static_polygon_filenames: static_polygons.add(_FeatureCollection(stat)) else: for file in url[0]: rotation_filenames.append(_collect_file_extension(download_from_web(file, verbose, model_name=self.file_collection), [".rot"])) rotation_model = _RotationModel(rotation_filenames) for file in url[1]: topology_filenames.append(_collect_file_extension(download_from_web(file, verbose, model_name=self.file_collection), [".gpml"])) for file in topology_filenames: topology_features.add(_FeatureCollection(file)) for file in url[2]: static_polygon_filenames.append( _check_gpml_or_shp( _str_in_folder( _str_in_filename(download_from_web(url[0], verbose, model_name=self.file_collection), strings_to_include=DataCollection.static_polygon_strings_to_include(self) ), strings_to_ignore=DataCollection.static_polygon_strings_to_ignore(self) ) ) ) for stat in static_polygon_filenames: static_polygons.add(_FeatureCollection(stat)) break if found_collection is False: raise ValueError("{} is not in GPlately's DataServer.".format(self.file_collection)) if not rotation_filenames: print("No .rot files in {}. No rotation model created.".format(self.file_collection)) rotation_model = [] if not topology_filenames: print("No topology features in {}. No FeatureCollection created - unable to plot trenches, ridges and transforms.".format(self.file_collection)) topology_features = [] if not static_polygons: print("No static polygons in {}.".format(self.file_collection)) static_polygons = [] # add identifier for setting up DownloadServer independently rotation_model.reconstruction_identifier = self.file_collection return rotation_model, topology_features, static_polygons def get_topology_geometries(self): """Uses Pooch to download coastline, continent and COB (continent-ocean boundary) Shapely geometries from the requested plate model. These are needed to call the `PlotTopologies` object and visualise topological plates through time. Parameters ---------- verbose : bool, default True Toggle print messages regarding server/internet connection status, file availability etc. Returns ------- coastlines : instance of <pygplates.FeatureCollection> Present-day global coastline Shapely polylines cookie-cut using static polygons. Ready for reconstruction to a particular geological time and for plotting. continents : instance of <pygplates.FeatureCollection> Cookie-cutting Shapely polygons for non-oceanic regions (continents, inta-oceanic arcs, etc.) ready for reconstruction to a particular geological time and for plotting. COBs : instance of <pygplates.FeatureCollection> Shapely polylines resolved from .shp and/or .gpml topology files that represent the locations of the boundaries between oceanic and continental crust. Ready for reconstruction to a particular geological time and for plotting. Notes ----- This method accesses the plate reconstruction model ascribed to the `file_collection` string passed into the `DataServer` object. For example, if the object was called with `"Muller2019"`: gDownload = gplately.download.DataServer("Muller2019") coastlines, continents, COBs = gDownload.get_topology_geometries() the method will attempt to download `coastlines`, `continents` and `COBs` from the Müller et al. (2019) plate reconstruction model. If found, these files are returned as individual pyGPlates Feature Collections. They can be passed into: gPlot = gplately.plot.PlotTopologies(gplately.reconstruction.PlateReconstruction, time, continents, coastlines, COBs) to reconstruct features to a certain geological time. The `PlotTopologies` object provides simple methods to plot these geometries along with trenches, ridges and transforms (see documentation for more info). Note that the `PlateReconstruction` object is a parameter. * Note: If the requested plate model does not have a certain geometry, a message will be printed to alert the user. For example, if `get_topology_geometries()` is used with the `"Matthews2016"` plate model, the workflow will print the following message: No continent-ocean boundaries in Matthews2016. """ verbose = self.verbose # Locate all topology geometries from GPlately's DataCollection database = DataCollection.topology_geometries(self) coastlines = [] continents = [] COBs = [] # Find the requested plate model data collection found_collection = False for collection, url in database.items(): if self.file_collection.lower() == collection.lower(): found_collection = True if len(url) == 1: # Some plate models do not have reconstructable geometries i.e. Li et al. 2008 if url[0] is None: break else: fnames = _collection_sorter( download_from_web(url[0], verbose, model_name=self.file_collection), self.file_collection ) coastlines = _check_gpml_or_shp( _str_in_folder( _str_in_filename( fnames, strings_to_include=DataCollection.coastline_strings_to_include(self), strings_to_ignore=DataCollection.coastline_strings_to_ignore(self), file_collection=self.file_collection, file_collection_sensitive=False ), strings_to_ignore=DataCollection.coastline_strings_to_ignore(self) ) ) continents = _check_gpml_or_shp( _str_in_folder( _str_in_filename( fnames, strings_to_include=DataCollection.continent_strings_to_include(self), strings_to_ignore=DataCollection.continent_strings_to_ignore(self), file_collection=self.file_collection, file_collection_sensitive=False ), strings_to_ignore=DataCollection.continent_strings_to_ignore(self) ) ) COBs = _check_gpml_or_shp( _str_in_folder( _str_in_filename( fnames, strings_to_include=DataCollection.COB_strings_to_include(self), strings_to_ignore=DataCollection.COB_strings_to_ignore(self), file_collection=self.file_collection, file_collection_sensitive=False ), strings_to_ignore=DataCollection.COB_strings_to_ignore(self) ) ) else: for file in url[0]: if url[0] is not None: coastlines.append(_str_in_filename( download_from_web(file, verbose, model_name=self.file_collection), strings_to_include=["coastline"]) ) coastlines = _check_gpml_or_shp(coastlines) else: coastlines = [] for file in url[1]: if url[1] is not None: continents.append(_str_in_filename( download_from_web(file, verbose, model_name=self.file_collection), strings_to_include=["continent"]) ) continents = _check_gpml_or_shp(continents) else: continents = [] for file in url[2]: if url[2] is not None: COBs.append(_str_in_filename( download_from_web(file, verbose, model_name=self.file_collection), strings_to_include=["cob"]) ) COBs = _check_gpml_or_shp(COBs) else: COBs = [] break if found_collection is False: raise ValueError("{} is not in GPlately's DataServer.".format(self.file_collection)) if not coastlines: print("No coastlines in {}.".format(self.file_collection)) coastlines_featurecollection = [] else: #print(coastlines) coastlines_featurecollection = _FeatureCollection() for coastline in coastlines: coastlines_featurecollection.add(_FeatureCollection(coastline)) if not continents: print("No continents in {}.".format(self.file_collection)) continents_featurecollection = [] else: #print(continents) continents_featurecollection = _FeatureCollection() for continent in continents: continents_featurecollection.add(_FeatureCollection(continent)) if not COBs: print("No continent-ocean boundaries in {}.".format(self.file_collection)) COBs_featurecollection = [] else: #print(COBs) COBs_featurecollection = _FeatureCollection() for COB in COBs: COBs_featurecollection.add(_FeatureCollection(COB)) geometries = coastlines_featurecollection, continents_featurecollection, COBs_featurecollection return geometries def get_age_grid(self, time): """Downloads seafloor and paleo-age grids from the plate reconstruction model (`file_collection`) passed into the `DataServer` object. Stores grids in the "gplately" cache. Currently, `DataServer` supports the following age grids: * __Muller et al. 2019__ * `file_collection` = `Muller2019` * Time range: 0-250 Ma * Seafloor age grid rasters in netCDF format. * __Muller et al. 2016__ * `file_collection` = `Muller2016` * Time range: 0-240 Ma * Seafloor age grid rasters in netCDF format. * __Seton et al. 2012__ * `file_collection` = `Seton2012` * Time range: 0-200 Ma * Paleo-age grid rasters in netCDF format. Parameters ---------- time : int, or list of int, default=None Request an age grid from one (an integer) or multiple reconstruction times (a list of integers). Returns ------- a gplately.Raster object A gplately.Raster object containing the age grid. The age grid data can be extracted into a numpy ndarray or MaskedArray by appending `.data` to the variable assigned to `get_age_grid()`. For example: gdownload = gplately.DataServer("Muller2019") graster = gdownload.get_age_grid(time=100) graster_data = graster.data where `graster_data` is a numpy ndarray. Raises ----- ValueError If `time` (a single integer, or a list of integers representing reconstruction times to extract the age grids from) is not passed. Notes ----- The first time that `get_age_grid` is called for a specific time(s), the age grid(s) will be downloaded into the GPlately cache once. Upon successive calls of `get_age_grid` for the same reconstruction time(s), the age grids will not be re-downloaded; rather, they are re-accessed from the same cache provided the age grid(s) have not been moved or deleted. Examples -------- if the `DataServer` object was called with the `Muller2019` `file_collection` string: gDownload = gplately.download.DataServer("Muller2019") `get_age_grid` will download seafloor age grids from the Müller et al. (2019) plate reconstruction model for the geological time(s) requested in the `time` parameter. If found, these age grids are returned as masked arrays. For example, to download Müller et al. (2019) seafloor age grids for 0Ma, 1Ma and 100 Ma: age_grids = gDownload.get_age_grid([0, 1, 100]) """ age_grids = [] age_grid_links = DataCollection.netcdf4_age_grids(self, time) if not isinstance(time, list): time = [time] # For a single time passed that isn't in the valid time range, if not age_grid_links: raise ValueError( "{} {}Ma age grids are not on GPlately's DataServer.".format( self.file_collection, time[0] ) ) # For a list of times passed... for i, link in enumerate(age_grid_links): if not link: raise ValueError( "{} {}Ma age grids are not on GPlately's DataServer.".format( self.file_collection, time[i] ) ) age_grid_file = download_from_web( link, verbose=self.verbose, model_name=self.file_collection ) age_grid = _gplately.grids.Raster(data=age_grid_file) age_grids.append(age_grid) # One last check to alert user if the masked array grids were not processed properly if not age_grids: raise ValueError("{} netCDF4 age grids not found.".format(self.file_collection)) if len(age_grids) == 1: return age_grids[0] else: return age_grids def get_spreading_rate_grid(self, time): """Downloads seafloor spreading rate grids from the plate reconstruction model (`file_collection`) passed into the `DataServer` object. Stores grids in the "gplately" cache. Currently, `DataServer` supports spreading rate grids from the following plate models: * __Clennett et al. 2020__ * `file_collection` = `Clennett2020` * Time range: 0-250 Ma * Seafloor spreading rate grids in netCDF format. Parameters ---------- time : int, or list of int, default=None Request a spreading grid from one (an integer) or multiple reconstruction times (a list of integers). Returns ------- a gplately.Raster object A gplately.Raster object containing the spreading rate grid. The spreading rate grid data can be extracted into a numpy ndarray or MaskedArray by appending `.data` to the variable assigned to `get_spreading_rate_grid()`. For example: gdownload = gplately.DataServer("Clennett2020") graster = gdownload.get_spreading_rate_grid(time=100) graster_data = graster.data where `graster_data` is a numpy ndarray. Raises ----- ValueError If `time` (a single integer, or a list of integers representing reconstruction times to extract the spreading rate grids from) is not passed. Notes ----- The first time that `get_spreading_rate_grid` is called for a specific time(s), the spreading rate grid(s) will be downloaded into the GPlately cache once. Upon successive calls of `get_spreading_rate_grid` for the same reconstruction time(s), the grids will not be re-downloaded; rather, they are re-accessed from the same cache location provided they have not been moved or deleted. Examples -------- if the `DataServer` object was called with the `Clennett2020` `file_collection` string: gDownload = gplately.download.DataServer("Clennett2020") `get_spreading_rate_grid` will download seafloor spreading rate grids from the Clennett et al. (2020) plate reconstruction model for the geological time(s) requested in the `time` parameter. When found, these spreading rate grids are returned as masked arrays. For example, to download Clennett et al. (2020) seafloor spreading rate grids for 0Ma, 1Ma and 100 Ma as MaskedArray objects: spreading_rate_grids = gDownload.get_spreading_rate_grid([0, 1, 100]) """ spreading_rate_grids = [] spreading_rate_grid_links = DataCollection.netcdf4_spreading_rate_grids(self, time) if not isinstance(time, list): time = [time] # For a single time passed that isn't in the valid time range, if not spreading_rate_grid_links: raise ValueError( "{} {}Ma spreading rate grids are not on GPlately's DataServer.".format( self.file_collection, time[0] ) ) # For a list of times passed... for i, link in enumerate(spreading_rate_grid_links): if not link: raise ValueError( "{} {}Ma spreading rate grids are not on GPlately's DataServer.".format( self.file_collection, time[i] ) ) spreading_rate_grid_file = download_from_web( link, verbose=self.verbose, model_name=self.file_collection ) spreading_rate_grid = _gplately.grids.Raster(data=spreading_rate_grid_file) spreading_rate_grids.append(spreading_rate_grid) # One last check to alert user if the masked array grids were not processed properly if not spreading_rate_grids: raise ValueError("{} netCDF4 seafloor spreading rate grids not found.".format(self.file_collection)) if len(spreading_rate_grids) == 1: return spreading_rate_grids[0] else: return spreading_rate_grids def get_valid_times(self): """Returns a tuple of the valid plate model time range, (min_time, max_time). """ all_model_valid_times = DataCollection.plate_model_valid_reconstruction_times(self) min_time = None max_time = None for plate_model_name, valid_times in list(all_model_valid_times.items()): if plate_model_name.lower() == self.file_collection.lower(): min_time = valid_times[0] max_time = valid_times[1] if not min_time and not max_time: raise ValueError("Could not find the valid reconstruction time of {}".format(self.file_collection)) return (min_time, max_time) def get_raster(self, raster_id_string=None): """Downloads assorted raster data that are not associated with the plate reconstruction models supported by GPlately's `DataServer`. Stores rasters in the "gplately" cache. Currently, `DataServer` supports the following rasters and images: * __[ETOPO1](https://www.ngdc.noaa.gov/mgg/global/)__: * Filetypes available : TIF, netCDF (GRD) * `raster_id_string` = `"ETOPO1_grd"`, `"ETOPO1_tif"` (depending on the requested format) * A 1-arc minute global relief model combining lang topography and ocean bathymetry. * Citation: doi:10.7289/V5C8276M Parameters ---------- raster_id_string : str, default=None A string to identify which raster to download. Returns ------- a gplately.Raster object A gplately.Raster object containing the raster data. The gridded data can be extracted into a numpy ndarray or MaskedArray by appending `.data` to the variable assigned to `get_raster()`. For example: gdownload = gplately.DataServer("Muller2019") graster = gdownload.get_raster(raster_id_string, verbose) graster_data = graster.data where `graster_data` is a numpy ndarray. This array can be visualised using `matplotlib.pyplot.imshow` on a `cartopy.mpl.GeoAxis` GeoAxesSubplot (see example below). Raises ------ ValueError * if a `raster_id_string` is not supplied. Notes ----- Rasters obtained by this method are (so far) only reconstructed to present-day. Examples -------- To download ETOPO1 and plot it on a Mollweide projection: import gplately import numpy as np import matplotlib.pyplot as plt import cartopy.crs as ccrs gdownload = gplately.DataServer("Muller2019") etopo1 = gdownload.get_raster("ETOPO1_tif") fig = plt.figure(figsize=(18,14), dpi=300) ax = fig.add_subplot(111, projection=ccrs.Mollweide(central_longitude = -150)) ax2.imshow(etopo1, extent=[-180,180,-90,90], transform=ccrs.PlateCarree()) """ return get_raster(raster_id_string, self.verbose) def get_feature_data(self, feature_data_id_string=None): """Downloads assorted geological feature data from web servers (i.e. [GPlates 2.3 sample data](https://www.earthbyte.org/gplates-2-3-software-and-data-sets/)) into the "gplately" cache. Currently, `DataServer` supports the following feature data: * __Large igneous provinces from Johansson et al. (2018)__ Information ----------- * Formats: .gpmlz * `feature_data_id_string` = `Johansson2018` Citations --------- * Johansson, L., Zahirovic, S., and Müller, R. D., In Prep, The interplay between the eruption and weathering of Large Igneous Provinces and the deep-time carbon cycle: Geophysical Research Letters. - __Large igneous province products interpreted as plume products from Whittaker et al. (2015)__. Information ----------- * Formats: .gpmlz, .shp * `feature_data_id_string` = `Whittaker2015` Citations --------- * Whittaker, J. M., Afonso, J. C., Masterton, S., Müller, R. D., Wessel, P., Williams, S. E., & Seton, M. (2015). Long-term interaction between mid-ocean ridges and mantle plumes. Nature Geoscience, 8(6), 479-483. doi:10.1038/ngeo2437. - __Seafloor tectonic fabric (fracture zones, discordant zones, V-shaped structures, unclassified V-anomalies, propagating ridge lineations and extinct ridges) from Matthews et al. (2011)__ Information ----------- * Formats: .gpml * `feature_data_id_string` = `SeafloorFabric` Citations --------- * Matthews, K.J., Müller, R.D., Wessel, P. and Whittaker, J.M., 2011. The tectonic fabric of the ocean basins. Journal of Geophysical Research, 116(B12): B12109, DOI: 10.1029/2011JB008413. - __Present day surface hotspot/plume locations from Whittaker et al. (2013)__ Information ----------- * Formats: .gpmlz * `feature_data_id_string` = `Hotspots` Citation -------- * Whittaker, J., Afonso, J., Masterton, S., Müller, R., Wessel, P., Williams, S., and Seton, M., 2015, Long-term interaction between mid-ocean ridges and mantle plumes: Nature Geoscience, v. 8, no. 6, p. 479-483, doi:10.1038/ngeo2437. Parameters ---------- feature_data_id_string : str, default=None A string to identify which feature data to download to the cache (see list of supported feature data above). Returns ------- feature_data_filenames : instance of <pygplates.FeatureCollection>, or list of instance <pygplates.FeatureCollection> If a single set of feature data is downloaded, a single pyGPlates `FeatureCollection` object is returned. Otherwise, a list containing multiple pyGPlates `FeatureCollection` objects is returned (like for `SeafloorFabric`). In the latter case, feature reconstruction and plotting may have to be done iteratively. Raises ------ ValueError If a `feature_data_id_string` is not provided. Examples -------- For examples of plotting data downloaded with `get_feature_data`, see GPlately's sample notebook 05 - Working With Feature Geometries [here](https://github.com/GPlates/gplately/blob/master/Notebooks/05-WorkingWithFeatureGeometries.ipynb). """ if feature_data_id_string is None: raise ValueError( "Please specify which feature data to fetch." ) database = _gplately.data._feature_data() found_collection = False for collection, zip_url in database.items(): if feature_data_id_string.lower() == collection.lower(): found_collection = True feature_data_filenames = _collection_sorter( _collect_file_extension( download_from_web(zip_url[0], self.verbose), [".gpml", ".gpmlz"] ), collection ) break if found_collection is False: raise ValueError("{} are not in GPlately's DataServer.".format(feature_data_id_string)) feat_data = _FeatureCollection() if len(feature_data_filenames) == 1: feat_data.add(_FeatureCollection(feature_data_filenames[0])) return feat_data else: feat_data=[] for file in feature_data_filenames: feat_data.append(_FeatureCollection(file)) return feat_dataMethods
def get_age_grid(self, time)-
Downloads seafloor and paleo-age grids from the plate reconstruction model (
file_collection) passed into theDataServerobject. Stores grids in the "gplately" cache.Currently,
DataServersupports the following age grids:-
Muller et al. 2019
file_collection=Muller2019- Time range: 0-250 Ma
- Seafloor age grid rasters in netCDF format.
-
Muller et al. 2016
file_collection=Muller2016- Time range: 0-240 Ma
- Seafloor age grid rasters in netCDF format.
-
Seton et al. 2012
file_collection=Seton2012- Time range: 0-200 Ma
- Paleo-age grid rasters in netCDF format.
Parameters
time:int,orlistofint, default=None- Request an age grid from one (an integer) or multiple reconstruction times (a list of integers).
Returns
a Raster object-
A gplately.Raster object containing the age grid. The age grid data can be extracted into a numpy ndarray or MaskedArray by appending
.datato the variable assigned toget_age_grid().For example:
gdownload = gplately.DataServer("Muller2019") graster = gdownload.get_age_grid(time=100) graster_data = graster.datawhere
graster_datais a numpy ndarray.
Raises
ValueError- If
time(a single integer, or a list of integers representing reconstruction times to extract the age grids from) is not passed.
Notes
The first time that
get_age_gridis called for a specific time(s), the age grid(s) will be downloaded into the GPlately cache once. Upon successive calls ofget_age_gridfor the same reconstruction time(s), the age grids will not be re-downloaded; rather, they are re-accessed from the same cache provided the age grid(s) have not been moved or deleted.Examples
if the
DataServerobject was called with theMuller2019file_collectionstring:gDownload = gplately.download.DataServer("Muller2019")get_age_gridwill download seafloor age grids from the Müller et al. (2019) plate reconstruction model for the geological time(s) requested in thetimeparameter. If found, these age grids are returned as masked arrays.For example, to download Müller et al. (2019) seafloor age grids for 0Ma, 1Ma and 100 Ma:
age_grids = gDownload.get_age_grid([0, 1, 100])Expand source code
def get_age_grid(self, time): """Downloads seafloor and paleo-age grids from the plate reconstruction model (`file_collection`) passed into the `DataServer` object. Stores grids in the "gplately" cache. Currently, `DataServer` supports the following age grids: * __Muller et al. 2019__ * `file_collection` = `Muller2019` * Time range: 0-250 Ma * Seafloor age grid rasters in netCDF format. * __Muller et al. 2016__ * `file_collection` = `Muller2016` * Time range: 0-240 Ma * Seafloor age grid rasters in netCDF format. * __Seton et al. 2012__ * `file_collection` = `Seton2012` * Time range: 0-200 Ma * Paleo-age grid rasters in netCDF format. Parameters ---------- time : int, or list of int, default=None Request an age grid from one (an integer) or multiple reconstruction times (a list of integers). Returns ------- a gplately.Raster object A gplately.Raster object containing the age grid. The age grid data can be extracted into a numpy ndarray or MaskedArray by appending `.data` to the variable assigned to `get_age_grid()`. For example: gdownload = gplately.DataServer("Muller2019") graster = gdownload.get_age_grid(time=100) graster_data = graster.data where `graster_data` is a numpy ndarray. Raises ----- ValueError If `time` (a single integer, or a list of integers representing reconstruction times to extract the age grids from) is not passed. Notes ----- The first time that `get_age_grid` is called for a specific time(s), the age grid(s) will be downloaded into the GPlately cache once. Upon successive calls of `get_age_grid` for the same reconstruction time(s), the age grids will not be re-downloaded; rather, they are re-accessed from the same cache provided the age grid(s) have not been moved or deleted. Examples -------- if the `DataServer` object was called with the `Muller2019` `file_collection` string: gDownload = gplately.download.DataServer("Muller2019") `get_age_grid` will download seafloor age grids from the Müller et al. (2019) plate reconstruction model for the geological time(s) requested in the `time` parameter. If found, these age grids are returned as masked arrays. For example, to download Müller et al. (2019) seafloor age grids for 0Ma, 1Ma and 100 Ma: age_grids = gDownload.get_age_grid([0, 1, 100]) """ age_grids = [] age_grid_links = DataCollection.netcdf4_age_grids(self, time) if not isinstance(time, list): time = [time] # For a single time passed that isn't in the valid time range, if not age_grid_links: raise ValueError( "{} {}Ma age grids are not on GPlately's DataServer.".format( self.file_collection, time[0] ) ) # For a list of times passed... for i, link in enumerate(age_grid_links): if not link: raise ValueError( "{} {}Ma age grids are not on GPlately's DataServer.".format( self.file_collection, time[i] ) ) age_grid_file = download_from_web( link, verbose=self.verbose, model_name=self.file_collection ) age_grid = _gplately.grids.Raster(data=age_grid_file) age_grids.append(age_grid) # One last check to alert user if the masked array grids were not processed properly if not age_grids: raise ValueError("{} netCDF4 age grids not found.".format(self.file_collection)) if len(age_grids) == 1: return age_grids[0] else: return age_grids -
def get_feature_data(self, feature_data_id_string=None)-
Downloads assorted geological feature data from web servers (i.e. GPlates 2.3 sample data) into the "gplately" cache.
Currently,
DataServersupports the following feature data:-
Large igneous provinces from Johansson et al. (2018)
Information
- Formats: .gpmlz
feature_data_id_string=Johansson2018
Citations
- Johansson, L., Zahirovic, S., and Müller, R. D., In Prep, The interplay between the eruption and weathering of Large Igneous Provinces and the deep-time carbon cycle: Geophysical Research Letters.
-
Large igneous province products interpreted as plume products from Whittaker et al. (2015).
Information
- Formats: .gpmlz, .shp
feature_data_id_string=Whittaker2015
Citations
- Whittaker, J. M., Afonso, J. C., Masterton, S., Müller, R. D., Wessel, P., Williams, S. E., & Seton, M. (2015). Long-term interaction between mid-ocean ridges and mantle plumes. Nature Geoscience, 8(6), 479-483. doi:10.1038/ngeo2437.
-
Seafloor tectonic fabric (fracture zones, discordant zones, V-shaped structures, unclassified V-anomalies, propagating ridge lineations and extinct ridges) from Matthews et al. (2011)
Information
- Formats: .gpml
feature_data_id_string=SeafloorFabric
Citations
- Matthews, K.J., Müller, R.D., Wessel, P. and Whittaker, J.M., 2011. The tectonic fabric of the ocean basins. Journal of Geophysical Research, 116(B12): B12109, DOI: 10.1029/2011JB008413.
-
Present day surface hotspot/plume locations from Whittaker et al. (2013)
Information
- Formats: .gpmlz
feature_data_id_string=Hotspots
Citation
- Whittaker, J., Afonso, J., Masterton, S., Müller, R., Wessel, P., Williams, S., and Seton, M., 2015, Long-term interaction between mid-ocean ridges and mantle plumes: Nature Geoscience, v. 8, no. 6, p. 479-483, doi:10.1038/ngeo2437.
Parameters
feature_data_id_string:str, default=None- A string to identify which feature data to download to the cache (see list of supported feature data above).
Returns
feature_data_filenames:instanceof<pygplates.FeatureCollection>,orlistofinstance <pygplates.FeatureCollection>- If a single set of feature data is downloaded, a single pyGPlates
FeatureCollectionobject is returned. Otherwise, a list containing multiple pyGPlatesFeatureCollectionobjects is returned (like forSeafloorFabric). In the latter case, feature reconstruction and plotting may have to be done iteratively.
Raises
ValueError- If a
feature_data_id_stringis not provided.
Examples
For examples of plotting data downloaded with
get_feature_data, see GPlately's sample notebook 05 - Working With Feature Geometries here.Expand source code
def get_feature_data(self, feature_data_id_string=None): """Downloads assorted geological feature data from web servers (i.e. [GPlates 2.3 sample data](https://www.earthbyte.org/gplates-2-3-software-and-data-sets/)) into the "gplately" cache. Currently, `DataServer` supports the following feature data: * __Large igneous provinces from Johansson et al. (2018)__ Information ----------- * Formats: .gpmlz * `feature_data_id_string` = `Johansson2018` Citations --------- * Johansson, L., Zahirovic, S., and Müller, R. D., In Prep, The interplay between the eruption and weathering of Large Igneous Provinces and the deep-time carbon cycle: Geophysical Research Letters. - __Large igneous province products interpreted as plume products from Whittaker et al. (2015)__. Information ----------- * Formats: .gpmlz, .shp * `feature_data_id_string` = `Whittaker2015` Citations --------- * Whittaker, J. M., Afonso, J. C., Masterton, S., Müller, R. D., Wessel, P., Williams, S. E., & Seton, M. (2015). Long-term interaction between mid-ocean ridges and mantle plumes. Nature Geoscience, 8(6), 479-483. doi:10.1038/ngeo2437. - __Seafloor tectonic fabric (fracture zones, discordant zones, V-shaped structures, unclassified V-anomalies, propagating ridge lineations and extinct ridges) from Matthews et al. (2011)__ Information ----------- * Formats: .gpml * `feature_data_id_string` = `SeafloorFabric` Citations --------- * Matthews, K.J., Müller, R.D., Wessel, P. and Whittaker, J.M., 2011. The tectonic fabric of the ocean basins. Journal of Geophysical Research, 116(B12): B12109, DOI: 10.1029/2011JB008413. - __Present day surface hotspot/plume locations from Whittaker et al. (2013)__ Information ----------- * Formats: .gpmlz * `feature_data_id_string` = `Hotspots` Citation -------- * Whittaker, J., Afonso, J., Masterton, S., Müller, R., Wessel, P., Williams, S., and Seton, M., 2015, Long-term interaction between mid-ocean ridges and mantle plumes: Nature Geoscience, v. 8, no. 6, p. 479-483, doi:10.1038/ngeo2437. Parameters ---------- feature_data_id_string : str, default=None A string to identify which feature data to download to the cache (see list of supported feature data above). Returns ------- feature_data_filenames : instance of <pygplates.FeatureCollection>, or list of instance <pygplates.FeatureCollection> If a single set of feature data is downloaded, a single pyGPlates `FeatureCollection` object is returned. Otherwise, a list containing multiple pyGPlates `FeatureCollection` objects is returned (like for `SeafloorFabric`). In the latter case, feature reconstruction and plotting may have to be done iteratively. Raises ------ ValueError If a `feature_data_id_string` is not provided. Examples -------- For examples of plotting data downloaded with `get_feature_data`, see GPlately's sample notebook 05 - Working With Feature Geometries [here](https://github.com/GPlates/gplately/blob/master/Notebooks/05-WorkingWithFeatureGeometries.ipynb). """ if feature_data_id_string is None: raise ValueError( "Please specify which feature data to fetch." ) database = _gplately.data._feature_data() found_collection = False for collection, zip_url in database.items(): if feature_data_id_string.lower() == collection.lower(): found_collection = True feature_data_filenames = _collection_sorter( _collect_file_extension( download_from_web(zip_url[0], self.verbose), [".gpml", ".gpmlz"] ), collection ) break if found_collection is False: raise ValueError("{} are not in GPlately's DataServer.".format(feature_data_id_string)) feat_data = _FeatureCollection() if len(feature_data_filenames) == 1: feat_data.add(_FeatureCollection(feature_data_filenames[0])) return feat_data else: feat_data=[] for file in feature_data_filenames: feat_data.append(_FeatureCollection(file)) return feat_data -
def get_plate_reconstruction_files(self)-
Downloads and constructs a
rotation model, a set oftopology_featuresand and a set ofstatic_polygonsneeded to call thePlateReconstructionobject.Returns
rotation_model:instanceof<pygplates.RotationModel>- A rotation model to query equivalent and/or relative topological plate rotations from a time in the past relative to another time in the past or to present day.
topology_features:instanceof<pygplates.FeatureCollection>- Point, polyline and/or polygon feature data that are reconstructable through geological time.
static_polygons:instanceof<pygplates.FeatureCollection>- Present-day polygons whose shapes do not change through geological time. They are used to cookie-cut dynamic polygons into identifiable topological plates (assigned an ID) according to their present-day locations.
Notes
This method accesses the plate reconstruction model ascribed to the
file_collectionstring passed into theDataServerobject. For example, if the object was called with"Muller2019":gDownload = gplately.download.DataServer("Muller2019") rotation_model, topology_features, static_polygons = gDownload.get_plate_reconstruction_files()the method will download a
rotation_model,topology_featuresandstatic_polygonsfrom the Müller et al. (2019) plate reconstruction model. Once the reconstruction objects are returned, they can be passed into:model = gplately.reconstruction.PlateReconstruction(rotation_model, topology_features, static_polygons)- Note: If the requested plate model does not have a certain file(s), a message will be printed
to alert the user. For example, using
get_plate_reconstruction_files()for the Torsvik and Cocks (2017) plate reconstruction model yields the printed message:No topology features in TorsvikCocks2017. No FeatureCollection created - unable to plot trenches, ridges and transforms. No continent-ocean boundaries in TorsvikCocks2017.
Expand source code
def get_plate_reconstruction_files(self): """Downloads and constructs a `rotation model`, a set of `topology_features` and and a set of `static_polygons` needed to call the `PlateReconstruction` object. Returns ------- rotation_model : instance of <pygplates.RotationModel> A rotation model to query equivalent and/or relative topological plate rotations from a time in the past relative to another time in the past or to present day. topology_features : instance of <pygplates.FeatureCollection> Point, polyline and/or polygon feature data that are reconstructable through geological time. static_polygons : instance of <pygplates.FeatureCollection> Present-day polygons whose shapes do not change through geological time. They are used to cookie-cut dynamic polygons into identifiable topological plates (assigned an ID) according to their present-day locations. Notes ----- This method accesses the plate reconstruction model ascribed to the `file_collection` string passed into the `DataServer` object. For example, if the object was called with `"Muller2019"`: gDownload = gplately.download.DataServer("Muller2019") rotation_model, topology_features, static_polygons = gDownload.get_plate_reconstruction_files() the method will download a `rotation_model`, `topology_features` and `static_polygons` from the Müller et al. (2019) plate reconstruction model. Once the reconstruction objects are returned, they can be passed into: model = gplately.reconstruction.PlateReconstruction(rotation_model, topology_features, static_polygons) * Note: If the requested plate model does not have a certain file(s), a message will be printed to alert the user. For example, using `get_plate_reconstruction_files()` for the Torsvik and Cocks (2017) plate reconstruction model yields the printed message: No topology features in TorsvikCocks2017. No FeatureCollection created - unable to plot trenches, ridges and transforms. No continent-ocean boundaries in TorsvikCocks2017. """ verbose = self.verbose rotation_filenames = [] rotation_model = [] topology_filenames = [] topology_features = _FeatureCollection() static_polygons= _FeatureCollection() static_polygon_filenames = [] # Locate all plate reconstruction files from GPlately's DataCollection database = DataCollection.plate_reconstruction_files(self) # Set to true if we find the given collection in our database found_collection = False for collection, url in database.items(): # Only continue if the user's chosen collection exists in our database if self.file_collection.lower() == collection.lower(): found_collection = True if len(url) == 1: fnames = _collection_sorter( download_from_web(url[0], verbose, model_name=self.file_collection), self.file_collection ) rotation_filenames = _collect_file_extension( _str_in_folder( _str_in_filename(fnames, strings_to_include=DataCollection.rotation_strings_to_include(self), strings_to_ignore=DataCollection.rotation_strings_to_ignore(self), file_collection=self.file_collection, file_collection_sensitive=True ), strings_to_ignore=DataCollection.rotation_strings_to_ignore(self) ), [".rot"] ) #print(rotation_filenames) rotation_model = _RotationModel(rotation_filenames) topology_filenames = _collect_file_extension( _str_in_folder( _str_in_filename(fnames, strings_to_include=DataCollection.dynamic_polygon_strings_to_include(self), strings_to_ignore=DataCollection.dynamic_polygon_strings_to_ignore(self), file_collection=self.file_collection, file_collection_sensitive=False, ), strings_to_ignore=DataCollection.dynamic_polygon_strings_to_ignore(self) ), [".gpml", ".gpmlz"] ) #print(topology_filenames) for file in topology_filenames: topology_features.add(_FeatureCollection(file)) static_polygon_filenames = _check_gpml_or_shp( _str_in_folder( _str_in_filename(fnames, strings_to_include=DataCollection.static_polygon_strings_to_include(self), strings_to_ignore=DataCollection.static_polygon_strings_to_ignore(self), file_collection=self.file_collection, file_collection_sensitive=False ), strings_to_ignore=DataCollection.static_polygon_strings_to_ignore(self) ) ) #print(static_polygon_filenames) for stat in static_polygon_filenames: static_polygons.add(_FeatureCollection(stat)) else: for file in url[0]: rotation_filenames.append(_collect_file_extension(download_from_web(file, verbose, model_name=self.file_collection), [".rot"])) rotation_model = _RotationModel(rotation_filenames) for file in url[1]: topology_filenames.append(_collect_file_extension(download_from_web(file, verbose, model_name=self.file_collection), [".gpml"])) for file in topology_filenames: topology_features.add(_FeatureCollection(file)) for file in url[2]: static_polygon_filenames.append( _check_gpml_or_shp( _str_in_folder( _str_in_filename(download_from_web(url[0], verbose, model_name=self.file_collection), strings_to_include=DataCollection.static_polygon_strings_to_include(self) ), strings_to_ignore=DataCollection.static_polygon_strings_to_ignore(self) ) ) ) for stat in static_polygon_filenames: static_polygons.add(_FeatureCollection(stat)) break if found_collection is False: raise ValueError("{} is not in GPlately's DataServer.".format(self.file_collection)) if not rotation_filenames: print("No .rot files in {}. No rotation model created.".format(self.file_collection)) rotation_model = [] if not topology_filenames: print("No topology features in {}. No FeatureCollection created - unable to plot trenches, ridges and transforms.".format(self.file_collection)) topology_features = [] if not static_polygons: print("No static polygons in {}.".format(self.file_collection)) static_polygons = [] # add identifier for setting up DownloadServer independently rotation_model.reconstruction_identifier = self.file_collection return rotation_model, topology_features, static_polygons def get_raster(self, raster_id_string=None)-
Downloads assorted raster data that are not associated with the plate reconstruction models supported by GPlately's
DataServer. Stores rasters in the "gplately" cache.Currently,
DataServersupports the following rasters and images:- ETOPO1:
- Filetypes available : TIF, netCDF (GRD)
raster_id_string="ETOPO1_grd","ETOPO1_tif"(depending on the requested format)- A 1-arc minute global relief model combining lang topography and ocean bathymetry.
- Citation: doi:10.7289/V5C8276M
Parameters
raster_id_string:str, default=None- A string to identify which raster to download.
Returns
a Raster object-
A gplately.Raster object containing the raster data. The gridded data can be extracted into a numpy ndarray or MaskedArray by appending
.datato the variable assigned toget_raster().For example:
gdownload = gplately.DataServer("Muller2019") graster = gdownload.get_raster(raster_id_string, verbose) graster_data = graster.datawhere
graster_datais a numpy ndarray. This array can be visualised usingmatplotlib.pyplot.imshowon acartopy.mpl.GeoAxisGeoAxesSubplot (see example below).
Raises
ValueError-
- if a
raster_id_stringis not supplied.
- if a
Notes
Rasters obtained by this method are (so far) only reconstructed to present-day.
Examples
To download ETOPO1 and plot it on a Mollweide projection:
import gplately import numpy as np import matplotlib.pyplot as plt import cartopy.crs as ccrs gdownload = gplately.DataServer("Muller2019") etopo1 = gdownload.get_raster("ETOPO1_tif") fig = plt.figure(figsize=(18,14), dpi=300) ax = fig.add_subplot(111, projection=ccrs.Mollweide(central_longitude = -150)) ax2.imshow(etopo1, extent=[-180,180,-90,90], transform=ccrs.PlateCarree())Expand source code
def get_raster(self, raster_id_string=None): """Downloads assorted raster data that are not associated with the plate reconstruction models supported by GPlately's `DataServer`. Stores rasters in the "gplately" cache. Currently, `DataServer` supports the following rasters and images: * __[ETOPO1](https://www.ngdc.noaa.gov/mgg/global/)__: * Filetypes available : TIF, netCDF (GRD) * `raster_id_string` = `"ETOPO1_grd"`, `"ETOPO1_tif"` (depending on the requested format) * A 1-arc minute global relief model combining lang topography and ocean bathymetry. * Citation: doi:10.7289/V5C8276M Parameters ---------- raster_id_string : str, default=None A string to identify which raster to download. Returns ------- a gplately.Raster object A gplately.Raster object containing the raster data. The gridded data can be extracted into a numpy ndarray or MaskedArray by appending `.data` to the variable assigned to `get_raster()`. For example: gdownload = gplately.DataServer("Muller2019") graster = gdownload.get_raster(raster_id_string, verbose) graster_data = graster.data where `graster_data` is a numpy ndarray. This array can be visualised using `matplotlib.pyplot.imshow` on a `cartopy.mpl.GeoAxis` GeoAxesSubplot (see example below). Raises ------ ValueError * if a `raster_id_string` is not supplied. Notes ----- Rasters obtained by this method are (so far) only reconstructed to present-day. Examples -------- To download ETOPO1 and plot it on a Mollweide projection: import gplately import numpy as np import matplotlib.pyplot as plt import cartopy.crs as ccrs gdownload = gplately.DataServer("Muller2019") etopo1 = gdownload.get_raster("ETOPO1_tif") fig = plt.figure(figsize=(18,14), dpi=300) ax = fig.add_subplot(111, projection=ccrs.Mollweide(central_longitude = -150)) ax2.imshow(etopo1, extent=[-180,180,-90,90], transform=ccrs.PlateCarree()) """ return get_raster(raster_id_string, self.verbose) - ETOPO1:
def get_spreading_rate_grid(self, time)-
Downloads seafloor spreading rate grids from the plate reconstruction model (
file_collection) passed into theDataServerobject. Stores grids in the "gplately" cache.Currently,
DataServersupports spreading rate grids from the following plate models:-
Clennett et al. 2020
file_collection=Clennett2020- Time range: 0-250 Ma
- Seafloor spreading rate grids in netCDF format.
Parameters
time:int,orlistofint, default=None- Request a spreading grid from one (an integer) or multiple reconstruction times (a list of integers).
Returns
a Raster object-
A gplately.Raster object containing the spreading rate grid. The spreading rate grid data can be extracted into a numpy ndarray or MaskedArray by appending
.datato the variable assigned toget_spreading_rate_grid().For example:
gdownload = gplately.DataServer("Clennett2020") graster = gdownload.get_spreading_rate_grid(time=100) graster_data = graster.datawhere
graster_datais a numpy ndarray.
Raises
ValueError- If
time(a single integer, or a list of integers representing reconstruction times to extract the spreading rate grids from) is not passed.
Notes
The first time that
get_spreading_rate_gridis called for a specific time(s), the spreading rate grid(s) will be downloaded into the GPlately cache once. Upon successive calls ofget_spreading_rate_gridfor the same reconstruction time(s), the grids will not be re-downloaded; rather, they are re-accessed from the same cache location provided they have not been moved or deleted.Examples
if the
DataServerobject was called with theClennett2020file_collectionstring:gDownload = gplately.download.DataServer("Clennett2020")get_spreading_rate_gridwill download seafloor spreading rate grids from the Clennett et al. (2020) plate reconstruction model for the geological time(s) requested in thetimeparameter. When found, these spreading rate grids are returned as masked arrays.For example, to download Clennett et al. (2020) seafloor spreading rate grids for 0Ma, 1Ma and 100 Ma as MaskedArray objects:
spreading_rate_grids = gDownload.get_spreading_rate_grid([0, 1, 100])Expand source code
def get_spreading_rate_grid(self, time): """Downloads seafloor spreading rate grids from the plate reconstruction model (`file_collection`) passed into the `DataServer` object. Stores grids in the "gplately" cache. Currently, `DataServer` supports spreading rate grids from the following plate models: * __Clennett et al. 2020__ * `file_collection` = `Clennett2020` * Time range: 0-250 Ma * Seafloor spreading rate grids in netCDF format. Parameters ---------- time : int, or list of int, default=None Request a spreading grid from one (an integer) or multiple reconstruction times (a list of integers). Returns ------- a gplately.Raster object A gplately.Raster object containing the spreading rate grid. The spreading rate grid data can be extracted into a numpy ndarray or MaskedArray by appending `.data` to the variable assigned to `get_spreading_rate_grid()`. For example: gdownload = gplately.DataServer("Clennett2020") graster = gdownload.get_spreading_rate_grid(time=100) graster_data = graster.data where `graster_data` is a numpy ndarray. Raises ----- ValueError If `time` (a single integer, or a list of integers representing reconstruction times to extract the spreading rate grids from) is not passed. Notes ----- The first time that `get_spreading_rate_grid` is called for a specific time(s), the spreading rate grid(s) will be downloaded into the GPlately cache once. Upon successive calls of `get_spreading_rate_grid` for the same reconstruction time(s), the grids will not be re-downloaded; rather, they are re-accessed from the same cache location provided they have not been moved or deleted. Examples -------- if the `DataServer` object was called with the `Clennett2020` `file_collection` string: gDownload = gplately.download.DataServer("Clennett2020") `get_spreading_rate_grid` will download seafloor spreading rate grids from the Clennett et al. (2020) plate reconstruction model for the geological time(s) requested in the `time` parameter. When found, these spreading rate grids are returned as masked arrays. For example, to download Clennett et al. (2020) seafloor spreading rate grids for 0Ma, 1Ma and 100 Ma as MaskedArray objects: spreading_rate_grids = gDownload.get_spreading_rate_grid([0, 1, 100]) """ spreading_rate_grids = [] spreading_rate_grid_links = DataCollection.netcdf4_spreading_rate_grids(self, time) if not isinstance(time, list): time = [time] # For a single time passed that isn't in the valid time range, if not spreading_rate_grid_links: raise ValueError( "{} {}Ma spreading rate grids are not on GPlately's DataServer.".format( self.file_collection, time[0] ) ) # For a list of times passed... for i, link in enumerate(spreading_rate_grid_links): if not link: raise ValueError( "{} {}Ma spreading rate grids are not on GPlately's DataServer.".format( self.file_collection, time[i] ) ) spreading_rate_grid_file = download_from_web( link, verbose=self.verbose, model_name=self.file_collection ) spreading_rate_grid = _gplately.grids.Raster(data=spreading_rate_grid_file) spreading_rate_grids.append(spreading_rate_grid) # One last check to alert user if the masked array grids were not processed properly if not spreading_rate_grids: raise ValueError("{} netCDF4 seafloor spreading rate grids not found.".format(self.file_collection)) if len(spreading_rate_grids) == 1: return spreading_rate_grids[0] else: return spreading_rate_grids -
def get_topology_geometries(self)-
Uses Pooch to download coastline, continent and COB (continent-ocean boundary) Shapely geometries from the requested plate model. These are needed to call the
PlotTopologiesobject and visualise topological plates through time.Parameters
verbose:bool, defaultTrue- Toggle print messages regarding server/internet connection status, file availability etc.
Returns
coastlines:instanceof<pygplates.FeatureCollection>- Present-day global coastline Shapely polylines cookie-cut using static polygons. Ready for reconstruction to a particular geological time and for plotting.
continents:instanceof<pygplates.FeatureCollection>- Cookie-cutting Shapely polygons for non-oceanic regions (continents, inta-oceanic arcs, etc.) ready for reconstruction to a particular geological time and for plotting.
COBs:instanceof<pygplates.FeatureCollection>- Shapely polylines resolved from .shp and/or .gpml topology files that represent the locations of the boundaries between oceanic and continental crust. Ready for reconstruction to a particular geological time and for plotting.
Notes
This method accesses the plate reconstruction model ascribed to the
file_collectionstring passed into theDataServerobject. For example, if the object was called with"Muller2019":gDownload = gplately.download.DataServer("Muller2019") coastlines, continents, COBs = gDownload.get_topology_geometries()the method will attempt to download
coastlines,continentsandCOBsfrom the Müller et al. (2019) plate reconstruction model. If found, these files are returned as individual pyGPlates Feature Collections. They can be passed into:gPlot = gplately.plot.PlotTopologies(gplately.reconstruction.PlateReconstruction, time, continents, coastlines, COBs)to reconstruct features to a certain geological time. The
PlotTopologiesobject provides simple methods to plot these geometries along with trenches, ridges and transforms (see documentation for more info). Note that thePlateReconstructionobject is a parameter.- Note: If the requested plate model does not have a certain geometry, a
message will be printed to alert the user. For example, if
get_topology_geometries()is used with the"Matthews2016"plate model, the workflow will print the following message:No continent-ocean boundaries in Matthews2016.
Expand source code
def get_topology_geometries(self): """Uses Pooch to download coastline, continent and COB (continent-ocean boundary) Shapely geometries from the requested plate model. These are needed to call the `PlotTopologies` object and visualise topological plates through time. Parameters ---------- verbose : bool, default True Toggle print messages regarding server/internet connection status, file availability etc. Returns ------- coastlines : instance of <pygplates.FeatureCollection> Present-day global coastline Shapely polylines cookie-cut using static polygons. Ready for reconstruction to a particular geological time and for plotting. continents : instance of <pygplates.FeatureCollection> Cookie-cutting Shapely polygons for non-oceanic regions (continents, inta-oceanic arcs, etc.) ready for reconstruction to a particular geological time and for plotting. COBs : instance of <pygplates.FeatureCollection> Shapely polylines resolved from .shp and/or .gpml topology files that represent the locations of the boundaries between oceanic and continental crust. Ready for reconstruction to a particular geological time and for plotting. Notes ----- This method accesses the plate reconstruction model ascribed to the `file_collection` string passed into the `DataServer` object. For example, if the object was called with `"Muller2019"`: gDownload = gplately.download.DataServer("Muller2019") coastlines, continents, COBs = gDownload.get_topology_geometries() the method will attempt to download `coastlines`, `continents` and `COBs` from the Müller et al. (2019) plate reconstruction model. If found, these files are returned as individual pyGPlates Feature Collections. They can be passed into: gPlot = gplately.plot.PlotTopologies(gplately.reconstruction.PlateReconstruction, time, continents, coastlines, COBs) to reconstruct features to a certain geological time. The `PlotTopologies` object provides simple methods to plot these geometries along with trenches, ridges and transforms (see documentation for more info). Note that the `PlateReconstruction` object is a parameter. * Note: If the requested plate model does not have a certain geometry, a message will be printed to alert the user. For example, if `get_topology_geometries()` is used with the `"Matthews2016"` plate model, the workflow will print the following message: No continent-ocean boundaries in Matthews2016. """ verbose = self.verbose # Locate all topology geometries from GPlately's DataCollection database = DataCollection.topology_geometries(self) coastlines = [] continents = [] COBs = [] # Find the requested plate model data collection found_collection = False for collection, url in database.items(): if self.file_collection.lower() == collection.lower(): found_collection = True if len(url) == 1: # Some plate models do not have reconstructable geometries i.e. Li et al. 2008 if url[0] is None: break else: fnames = _collection_sorter( download_from_web(url[0], verbose, model_name=self.file_collection), self.file_collection ) coastlines = _check_gpml_or_shp( _str_in_folder( _str_in_filename( fnames, strings_to_include=DataCollection.coastline_strings_to_include(self), strings_to_ignore=DataCollection.coastline_strings_to_ignore(self), file_collection=self.file_collection, file_collection_sensitive=False ), strings_to_ignore=DataCollection.coastline_strings_to_ignore(self) ) ) continents = _check_gpml_or_shp( _str_in_folder( _str_in_filename( fnames, strings_to_include=DataCollection.continent_strings_to_include(self), strings_to_ignore=DataCollection.continent_strings_to_ignore(self), file_collection=self.file_collection, file_collection_sensitive=False ), strings_to_ignore=DataCollection.continent_strings_to_ignore(self) ) ) COBs = _check_gpml_or_shp( _str_in_folder( _str_in_filename( fnames, strings_to_include=DataCollection.COB_strings_to_include(self), strings_to_ignore=DataCollection.COB_strings_to_ignore(self), file_collection=self.file_collection, file_collection_sensitive=False ), strings_to_ignore=DataCollection.COB_strings_to_ignore(self) ) ) else: for file in url[0]: if url[0] is not None: coastlines.append(_str_in_filename( download_from_web(file, verbose, model_name=self.file_collection), strings_to_include=["coastline"]) ) coastlines = _check_gpml_or_shp(coastlines) else: coastlines = [] for file in url[1]: if url[1] is not None: continents.append(_str_in_filename( download_from_web(file, verbose, model_name=self.file_collection), strings_to_include=["continent"]) ) continents = _check_gpml_or_shp(continents) else: continents = [] for file in url[2]: if url[2] is not None: COBs.append(_str_in_filename( download_from_web(file, verbose, model_name=self.file_collection), strings_to_include=["cob"]) ) COBs = _check_gpml_or_shp(COBs) else: COBs = [] break if found_collection is False: raise ValueError("{} is not in GPlately's DataServer.".format(self.file_collection)) if not coastlines: print("No coastlines in {}.".format(self.file_collection)) coastlines_featurecollection = [] else: #print(coastlines) coastlines_featurecollection = _FeatureCollection() for coastline in coastlines: coastlines_featurecollection.add(_FeatureCollection(coastline)) if not continents: print("No continents in {}.".format(self.file_collection)) continents_featurecollection = [] else: #print(continents) continents_featurecollection = _FeatureCollection() for continent in continents: continents_featurecollection.add(_FeatureCollection(continent)) if not COBs: print("No continent-ocean boundaries in {}.".format(self.file_collection)) COBs_featurecollection = [] else: #print(COBs) COBs_featurecollection = _FeatureCollection() for COB in COBs: COBs_featurecollection.add(_FeatureCollection(COB)) geometries = coastlines_featurecollection, continents_featurecollection, COBs_featurecollection return geometries def get_valid_times(self)-
Returns a tuple of the valid plate model time range, (min_time, max_time).
Expand source code
def get_valid_times(self): """Returns a tuple of the valid plate model time range, (min_time, max_time). """ all_model_valid_times = DataCollection.plate_model_valid_reconstruction_times(self) min_time = None max_time = None for plate_model_name, valid_times in list(all_model_valid_times.items()): if plate_model_name.lower() == self.file_collection.lower(): min_time = valid_times[0] max_time = valid_times[1] if not min_time and not max_time: raise ValueError("Could not find the valid reconstruction time of {}".format(self.file_collection)) return (min_time, max_time)
-
class PlateReconstruction (rotation_model, topology_features=None, static_polygons=None, anchor_plate_id=0)-
The
PlateReconstructionclass contains methods to reconstruct topology features to specific geological times given arotation_model, a set oftopology_featuresand a set ofstatic_polygons. Topological plate velocity data at specific geological times can also be calculated from these reconstructed features.Attributes
rotation_model:str,orinstanceof<pygplates.FeatureCollection>,or<pygplates.Feature>,orsequenceof<pygplates.Feature>,orinstanceof<pygplates.RotationModel>, defaultNone- A rotation model to query equivalent and/or relative topological plate rotations from a time in the past relative to another time in the past or to present day. Can be provided as a rotation filename, or rotation feature collection, or rotation feature, or sequence of rotation features, or a sequence (eg, a list or tuple) of any combination of those four types.
topology_features:str,ora sequence (eg,listortuple)ofinstancesof<pygplates.Feature>,ora single instanceof<pygplates.Feature>,oran instanceof<pygplates.FeatureCollection>, defaultNone- Reconstructable topological features like trenches, ridges and transforms. Can be provided as an optional topology-feature filename, or sequence of features, or a single feature.
static_polygons:str,orinstanceof<pygplates.Feature>,orsequenceof<pygplates.Feature>,or an instanceof<pygplates.FeatureCollection>, defaultNone- Present-day polygons whose shapes do not change through geological time. They are used to cookie-cut dynamic polygons into identifiable topological plates (assigned an ID) according to their present-day locations. Can be provided as a static polygon feature collection, or optional filename, or a single feature, or a sequence of features.
Expand source code
class PlateReconstruction(object): """The `PlateReconstruction` class contains methods to reconstruct topology features to specific geological times given a `rotation_model`, a set of `topology_features` and a set of `static_polygons`. Topological plate velocity data at specific geological times can also be calculated from these reconstructed features. Attributes ---------- rotation_model : str, or instance of <pygplates.FeatureCollection>, or <pygplates.Feature>, or sequence of <pygplates.Feature>, or instance of <pygplates.RotationModel>, default None A rotation model to query equivalent and/or relative topological plate rotations from a time in the past relative to another time in the past or to present day. Can be provided as a rotation filename, or rotation feature collection, or rotation feature, or sequence of rotation features, or a sequence (eg, a list or tuple) of any combination of those four types. topology_features : str, or a sequence (eg, `list` or `tuple`) of instances of <pygplates.Feature>, or a single instance of <pygplates.Feature>, or an instance of <pygplates.FeatureCollection>, default None Reconstructable topological features like trenches, ridges and transforms. Can be provided as an optional topology-feature filename, or sequence of features, or a single feature. static_polygons : str, or instance of <pygplates.Feature>, or sequence of <pygplates.Feature>,or an instance of <pygplates.FeatureCollection>, default None Present-day polygons whose shapes do not change through geological time. They are used to cookie-cut dynamic polygons into identifiable topological plates (assigned an ID) according to their present-day locations. Can be provided as a static polygon feature collection, or optional filename, or a single feature, or a sequence of features. """ def __init__( self, rotation_model, topology_features=None, static_polygons=None, anchor_plate_id=0, ): if hasattr(rotation_model, "reconstruction_identifier"): self.name = rotation_model.reconstruction_identifier else: self.name = None self.anchor_plate_id = int(anchor_plate_id) self.rotation_model = _RotationModel( rotation_model, default_anchor_plate_id=anchor_plate_id ) self.topology_features = _load_FeatureCollection(topology_features) self.static_polygons = _load_FeatureCollection(static_polygons) def __getstate__(self): filenames = { "rotation_model": self.rotation_model.filenames, "anchor_plate_id": self.anchor_plate_id, } if self.topology_features: filenames["topology_features"] = self.topology_features.filenames if self.static_polygons: filenames["static_polygons"] = self.static_polygons.filenames # # remove unpicklable items # del self.rotation_model, self.topology_features, self.static_polygons # # really make sure they're gone # self.rotation_model = None # self.topology_features = None # self.static_polygons = None return filenames def __setstate__(self, state): # reinstate unpicklable items self.rotation_model = _RotationModel( state["rotation_model"], default_anchor_plate_id=state["anchor_plate_id"] ) self.anchor_plate_id = state["anchor_plate_id"] self.topology_features = None self.static_polygons = None if "topology_features" in state: self.topology_features = _FeatureCollection() for topology in state["topology_features"]: self.topology_features.add(_FeatureCollection(topology)) if "static_polygons" in state: self.static_polygons = _FeatureCollection() for polygon in state["static_polygons"]: self.static_polygons.add(_FeatureCollection(polygon)) def tessellate_subduction_zones( self, time, tessellation_threshold_radians=0.001, ignore_warnings=False, return_geodataframe=False, **kwargs ): """Samples points along subduction zone trenches and obtains subduction data at a particular geological time. Resolves topologies at `time`, tessellates all resolved subducting features to within 'tessellation_threshold_radians' radians and obtains the following information for each sampled point along a trench: `tessellate_subduction_zones` returns a list of 10 vertically-stacked tuples with the following data per sampled trench point: * Col. 0 - longitude of sampled trench point * Col. 1 - latitude of sampled trench point * Col. 2 - subducting convergence (relative to trench) velocity magnitude (in cm/yr) * Col. 3 - subducting convergence velocity obliquity angle (angle between trench normal vector and convergence velocity vector) * Col. 4 - trench absolute (relative to anchor plate) velocity magnitude (in cm/yr) * Col. 5 - trench absolute velocity obliquity angle (angle between trench normal vector and trench absolute velocity vector) * Col. 6 - length of arc segment (in degrees) that current point is on * Col. 7 - trench normal azimuth angle (clockwise starting at North, ie, 0 to 360 degrees) at current point * Col. 8 - subducting plate ID * Col. 9 - trench plate ID Parameters ---------- time : float The reconstruction time (Ma) at which to query subduction convergence. tessellation_threshold_radians : float, default=0.001 The threshold sampling distance along the subducting trench (in radians). Returns ------- subduction_data : a list of vertically-stacked tuples The results for all tessellated points sampled along the trench. The size of the returned list is equal to the number of tessellated points. Each tuple in the list corresponds to a tessellated point and has the following tuple items: * Col. 0 - longitude of sampled trench point * Col. 1 - latitude of sampled trench point * Col. 2 - subducting convergence (relative to trench) velocity magnitude (in cm/yr) * Col. 3 - subducting convergence velocity obliquity angle (angle between trench normal vector and convergence velocity vector) * Col. 4 - trench absolute (relative to anchor plate) velocity magnitude (in cm/yr) * Col. 5 - trench absolute velocity obliquity angle (angle between trench normal vector and trench absolute velocity vector) * Col. 6 - length of arc segment (in degrees) that current point is on * Col. 7 - trench normal azimuth angle (clockwise starting at North, ie, 0 to 360 degrees) at current point * Col. 8 - subducting plate ID * Col. 9 - trench plate ID Notes ----- Each sampled point in the output is the midpoint of a great circle arc between two adjacent points in the trench polyline. The trench normal vector used in the obliquity calculations is perpendicular to the great circle arc of each point (arc midpoint) and pointing towards the overriding plate (rather than away from it). Each trench is sampled at approximately uniform intervals along its length (specified via a threshold sampling distance). The sampling along the entire length of a trench is not exactly uniform. Each segment along a trench is sampled such that the samples have a uniform spacing that is less than or equal to the threshold sampling distance. However each segment in a trench might have a slightly different spacing distance (since segment lengths are not integer multiples of the threshold sampling distance). The trench normal (at each arc segment mid-point) always points *towards* the overriding plate. The obliquity angles are in the range (-180 180). The range (0, 180) goes clockwise (when viewed from above the Earth) from the trench normal direction to the velocity vector. The range (0, -180) goes counter-clockwise. You can change the range (-180, 180) to the range (0, 360) by adding 360 to negative angles. The trench normal is perpendicular to the trench and pointing toward the overriding plate. Note that the convergence velocity magnitude is negative if the plates are diverging (if convergence obliquity angle is greater than 90 or less than -90). And note that the absolute velocity magnitude is negative if the trench (subduction zone) is moving towards the overriding plate (if absolute obliquity angle is less than 90 or greater than -90) - note that this ignores the kinematics of the subducting plate. The delta time interval used for velocity calculations is, by default, assumed to be 1Ma. """ from . import ptt as _ptt anchor_plate_id = kwargs.pop("anchor_plate_id", self.anchor_plate_id) if ignore_warnings: with warnings.catch_warnings(): warnings.simplefilter("ignore") subduction_data = _ptt.subduction_convergence.subduction_convergence( self.rotation_model, self.topology_features, tessellation_threshold_radians, float(time), anchor_plate_id=anchor_plate_id, **kwargs ) else: subduction_data = _ptt.subduction_convergence.subduction_convergence( self.rotation_model, self.topology_features, tessellation_threshold_radians, float(time), anchor_plate_id=anchor_plate_id, **kwargs ) subduction_data = np.vstack(subduction_data) if return_geodataframe: import geopandas as gpd from shapely import geometry coords = [ geometry.Point(lon, lat) for lon, lat in zip(subduction_data[:, 0], subduction_data[:, 1]) ] d = {"geometry": coords} labels = [ "convergence velocity (cm/yr)", "convergence obliquity angle (degrees)", "trench velocity (cm/yr)", "trench obliquity angle (degrees)", "length (degrees)", "trench normal angle (degrees)", "subducting plate ID", "overriding plate ID", ] for i, label in enumerate(labels): index = 2 + i d[label] = subduction_data[:, index] gdf = gpd.GeoDataFrame(d, geometry="geometry") return gdf else: return subduction_data def total_subduction_zone_length(self, time, use_ptt=False, ignore_warnings=False): """Calculates the total length of all mid-ocean ridges (km) at the specified geological time (Ma). if `use_ptt` is True Uses Plate Tectonic Tools' `subduction_convergence` module to calculate trench segment lengths on a unit sphere. The aggregated total subduction zone length is scaled to kilometres using the geocentric radius. Otherwise Resolves topology features ascribed to the `PlateReconstruction` model and extracts their shared boundary sections. The lengths of each trench boundary section are appended to the total subduction zone length. The total length is scaled to kilometres using a latitude-dependent (geocentric) Earth radius. Parameters ---------- time : int The geological time at which to calculate total mid-ocean ridge lengths. use_ptt : bool, default=False If set to `True`, the PTT method is used. ignore_warnings : bool, default=False Choose whether to ignore warning messages from PTT's `subduction_convergence` workflow. These warnings alert the user when certain subduction sub-segments are ignored - this happens when the trench segments have unidentifiable subduction polarities and/or subducting plates. Raises ------ ValueError If neither `use_pygplates` or `use_ptt` have been set to `True`. Returns ------- total_subduction_zone_length_kms : float The total subduction zone length (in km) at the specified `time`. """ from . import ptt as _ptt if use_ptt: with warnings.catch_warnings(): warnings.simplefilter("ignore") subduction_data = self.tessellate_subduction_zones( time, ignore_warnings=ignore_warnings ) trench_arcseg = subduction_data[:, 6] trench_pt_lat = subduction_data[:, 1] total_subduction_zone_length_kms = 0 for i, segment in enumerate(trench_arcseg): earth_radius = _tools.geocentric_radius(trench_pt_lat[i]) / 1e3 total_subduction_zone_length_kms += np.deg2rad(segment) * earth_radius return total_subduction_zone_length_kms else: resolved_topologies = [] shared_boundary_sections = [] pygplates.resolve_topologies( self.topology_features, self.rotation_model, resolved_topologies, time, shared_boundary_sections, ) total_subduction_zone_length_kms = 0.0 for shared_boundary_section in shared_boundary_sections: if ( shared_boundary_section.get_feature().get_feature_type() != pygplates.FeatureType.gpml_subduction_zone ): continue for ( shared_sub_segment ) in shared_boundary_section.get_shared_sub_segments(): clat, clon = ( shared_sub_segment.get_resolved_geometry() .get_centroid() .to_lat_lon() ) earth_radius = _tools.geocentric_radius(clat) / 1e3 total_subduction_zone_length_kms += ( shared_sub_segment.get_resolved_geometry().get_arc_length() * earth_radius ) return total_subduction_zone_length_kms def total_continental_arc_length( self, time, continental_grid, trench_arc_distance, ignore_warnings=True, ): """Calculates the total length of all global continental arcs (km) at the specified geological time (Ma). Uses Plate Tectonic Tools' `subduction_convergence` workflow to sample a given plate model's trench features into point features and obtain their subduction polarities. The resolved points are projected out by the `trench_arc_distance` and their new locations are linearly interpolated onto the supplied `continental_grid`. If the projected trench points lie in the grid, they are considered continental arc points, and their arc segment lengths are appended to the total continental arc length for the specified `time`. The total length is scaled to kilometres using the geocentric Earth radius. Parameters ---------- time : int The geological time at which to calculate total continental arc lengths. continental_grid: Raster, array_like, or str The continental grid used to identify continental arc points. Must be convertible to `Raster`. For an array, a global extent is assumed [-180,180,-90,90]. For a filename, the extent is obtained from the file. trench_arc_distance : float The trench-to-arc distance (in kilometres) to project sampled trench points out by in the direction of their subduction polarities. ignore_warnings : bool, default=True Choose whether to ignore warning messages from PTT's subduction_convergence workflow that alerts the user of subduction sub-segments that are ignored due to unidentified polarities and/or subducting plates. Returns ------- total_continental_arc_length_kms : float The continental arc length (in km) at the specified time. """ from . import grids as _grids if isinstance(continental_grid, _grids.Raster): graster = continental_grid elif isinstance(continental_grid, str): # Process the continental grid directory graster = _grids.Raster( data=continental_grid, realign=True, time=float(time), ) else: # Process the masked continental grid try: continental_grid = np.array(continental_grid) graster = _grids.Raster( data=continental_grid, extent=[-180, 180, -90, 90], time=float(time), ) except Exception as e: raise TypeError( "Invalid type for `continental_grid` (must be Raster," + " str, or array_like)" ) from e if (time != graster.time) and (not ignore_warnings): raise RuntimeWarning( "`continental_grid.time` ({}) ".format(graster.time) + "does not match `time` ({})".format(time) ) # Obtain trench data with Plate Tectonic Tools trench_data = self.tessellate_subduction_zones( time, ignore_warnings=ignore_warnings ) # Extract trench data trench_normal_azimuthal_angle = trench_data[:, 7] trench_arcseg = trench_data[:, 6] trench_pt_lon = trench_data[:, 0] trench_pt_lat = trench_data[:, 1] # Modify the trench-arc distance using the geocentric radius arc_distance = trench_arc_distance / ( _tools.geocentric_radius(trench_pt_lat) / 1000 ) # Project trench points out along trench-arc distance, and obtain their new lat-lon coordinates dlon = arc_distance * np.sin(np.radians(trench_normal_azimuthal_angle)) dlat = arc_distance * np.cos(np.radians(trench_normal_azimuthal_angle)) ilon = trench_pt_lon + np.degrees(dlon) ilat = trench_pt_lat + np.degrees(dlat) # Linearly interpolate projected points onto continental grids, and collect the indices of points that lie # within the grids. sampled_points = graster.interpolate( ilon, ilat, method="linear", return_indices=False, ) continental_indices = np.where(sampled_points > 0) point_lats = ilat[continental_indices] point_radii = _tools.geocentric_radius(point_lats) * 1.0e-3 # km segment_arclens = np.deg2rad(trench_arcseg[continental_indices]) segment_lengths = point_radii * segment_arclens return np.sum(segment_lengths) def tessellate_mid_ocean_ridges( self, time, tessellation_threshold_radians=0.001, ignore_warnings=False, return_geodataframe=False, **kwargs ): """Samples points along resolved spreading features (e.g. mid-ocean ridges) and calculates spreading rates and lengths of ridge segments at a particular geological time. Resolves topologies at `time`, tessellates all resolved spreading features to within 'tessellation_threshold_radians' radians. Returns a 4-column vertically stacked tuple with the following data. * Col. 0 - longitude of sampled ridge point * Col. 1 - latitude of sampled ridge point * Col. 2 - spreading velocity magnitude (in cm/yr) * Col. 3 - length of arc segment (in degrees) that current point is on All spreading feature types are considered. The transform segments of spreading features are ignored. Note: by default, the function assumes that a segment can deviate 45 degrees from the stage pole before it is considered a transform segment. Parameters ---------- time : float The reconstruction time (Ma) at which to query subduction convergence. tessellation_threshold_radians : float, default=0.001 The threshold sampling distance along the subducting trench (in radians). ignore_warnings : bool, default=False Choose to ignore warnings from Plate Tectonic Tools' ridge_spreading_rate workflow. Returns ------- ridge_data : a list of vertically-stacked tuples The results for all tessellated points sampled along the trench. The size of the returned list is equal to the number of tessellated points. Each tuple in the list corresponds to a tessellated point and has the following tuple items: * longitude of sampled point * latitude of sampled point * spreading velocity magnitude (in cm/yr) * length of arc segment (in degrees) that current point is on """ from . import ptt as _ptt anchor_plate_id = kwargs.pop("anchor_plate_id", self.anchor_plate_id) if ignore_warnings: with warnings.catch_warnings(): warnings.simplefilter("ignore") spreading_feature_types = [pygplates.FeatureType.gpml_mid_ocean_ridge] ridge_data = _ptt.ridge_spreading_rate.spreading_rates( self.rotation_model, self.topology_features, float(time), tessellation_threshold_radians, spreading_feature_types, anchor_plate_id=anchor_plate_id, **kwargs ) else: spreading_feature_types = [pygplates.FeatureType.gpml_mid_ocean_ridge] ridge_data = _ptt.ridge_spreading_rate.spreading_rates( self.rotation_model, self.topology_features, float(time), tessellation_threshold_radians, spreading_feature_types, anchor_plate_id=anchor_plate_id, **kwargs ) if not ridge_data: # the _ptt.ridge_spreading_rate.spreading_rates might return None return ridge_data = np.vstack(ridge_data) if return_geodataframe: import geopandas as gpd from shapely import geometry points = [ geometry.Point(lon, lat) for lon, lat in zip(ridge_data[:, 0], ridge_data[:, 1]) ] gdf = gpd.GeoDataFrame( { "geometry": points, "velocity (cm/yr)": ridge_data[:, 2], "length (degrees)": ridge_data[:, 3], }, geometry="geometry", ) return gdf else: return ridge_data def total_ridge_length(self, time, use_ptt=False, ignore_warnings=False): """Calculates the total length of all mid-ocean ridges (km) at the specified geological time (Ma). if `use_ptt` is True Uses Plate Tectonic Tools' `ridge_spreading_rate` workflow to calculate ridge segment lengths. Scales lengths to kilometres using the geocentric radius. Otherwise Resolves topology features of the PlateReconstruction model and extracts their shared boundary sections. The lengths of each GPML mid-ocean ridge shared boundary section are appended to the total ridge length. Scales lengths to kilometres using the geocentric radius. Parameters ---------- time : int The geological time at which to calculate total mid-ocean ridge lengths. use_ptt : bool, default=False If set to `True`, the PTT method is used. ignore_warnings : bool, default=False Choose whether to ignore warning messages from PTT's `ridge_spreading_rate` workflow. Raises ------ ValueError If neither `use_pygplates` or `use_ptt` have been set to `True`. Returns ------- total_ridge_length_kms : float The total length of global mid-ocean ridges (in kilometres) at the specified time. """ from . import ptt as _ptt if use_ptt is True: with warnings.catch_warnings(): warnings.simplefilter("ignore") ridge_data = self.tessellate_mid_ocean_ridges(time) ridge_arcseg = ridge_data[:, 3] ridge_pt_lat = ridge_data[:, 1] total_ridge_length_kms = 0 for i, segment in enumerate(ridge_arcseg): earth_radius = _tools.geocentric_radius(ridge_pt_lat[i]) / 1e3 total_ridge_length_kms += np.deg2rad(segment) * earth_radius return total_ridge_length_kms else: resolved_topologies = [] shared_boundary_sections = [] pygplates.resolve_topologies( self.topology_features, self.rotation_model, resolved_topologies, time, shared_boundary_sections, ) total_ridge_length_kms = 0.0 for shared_boundary_section in shared_boundary_sections: if ( shared_boundary_section.get_feature().get_feature_type() != pygplates.FeatureType.gpml_mid_ocean_ridge ): continue for ( shared_sub_segment ) in shared_boundary_section.get_shared_sub_segments(): clat, clon = ( shared_sub_segment.get_resolved_geometry() .get_centroid() .to_lat_lon() ) earth_radius = _tools.geocentric_radius(clat) / 1e3 total_ridge_length_kms += ( shared_sub_segment.get_resolved_geometry().get_arc_length() * earth_radius ) return total_ridge_length_kms def reconstruct( self, feature, to_time, from_time=0, anchor_plate_id=None, **kwargs ): """Reconstructs regular geological features, motion paths or flowlines to a specific geological time. Parameters ---------- feature : str, or instance of <pygplates.FeatureCollection>, or <pygplates.Feature>, or sequence of <pygplates.Feature> The geological features to reconstruct. Can be provided as a feature collection, or filename, or feature, or sequence of features, or a sequence (eg, a list or tuple) of any combination of those four types. to_time : float, or :class:`GeoTimeInstant` The specific geological time to reconstruct to. from_time : float, default=0 The specific geological time to reconstruct from. By default, this is set to present day. Raises `NotImplementedError` if `from_time` is not set to 0.0 Ma (present day). anchor_plate_id : int, default=None Reconstruct features with respect to a certain anchor plate. By default, reconstructions are made with respect to the absolute reference frame (anchor_plate_id = 0), like a stationary object in the mantle, unless otherwise specified. **reconstruct_type : ReconstructType, default=ReconstructType.feature_geometry The specific reconstruction type to generate based on input feature geometry type. Can be provided as pygplates.ReconstructType.feature_geometry to only reconstruct regular feature geometries, or pygplates.ReconstructType.motion_path to only reconstruct motion path features, or pygplates.ReconstructType.flowline to only reconstruct flowline features. Generates :class:`reconstructed feature geometries<ReconstructedFeatureGeometry>’, or :class:`reconstructed motion paths<ReconstructedMotionPath>’, or :class:`reconstructed flowlines<ReconstructedFlowline>’ respectively. **group_with_feature : bool, default=False Used to group reconstructed geometries with their features. This can be useful when a feature has more than one geometry and hence more than one reconstructed geometry. The output *reconstructed_geometries* then becomes a list of tuples where each tuple contains a :class:`feature<Feature>` and a ``list`` of reconstructed geometries. Note: this keyword argument only applies when *reconstructed_geometries* is a list because exported files are always grouped with their features. This is applicable to all ReconstructType features. **export_wrap_to_dateline : bool, default=True Wrap/clip reconstructed geometries to the dateline. Returns ------- reconstructed_features : list Reconstructed geological features (generated by the reconstruction) are appended to the list. The reconstructed geometries are output in the same order as that of their respective input features (in the parameter “features”). The order across input feature collections is also retained. This happens regardless of whether *features* and *reconstructed_features* include files or not. Note: if keyword argument group_with_feature=True then the list contains tuples that group each :class:`feature<Feature>` with a list of its reconstructed geometries. Raises ------ NotImplementedError if the starting time for reconstruction `from_time` is not equal to 0.0. """ from_time, to_time = float(from_time), float(to_time) reconstructed_features = [] if not anchor_plate_id: anchor_plate_id = self.anchor_plate_id pygplates.reconstruct( feature, self.rotation_model, reconstructed_features, to_time, anchor_plate_id=anchor_plate_id, **kwargs ) return reconstructed_features def get_point_velocities(self, lons, lats, time, delta_time=1.0): """Generates a velocity domain feature collection, resolves them into points, and calculates the north and east components of the velocity vector for each point in the domain at a particular geological `time`. Notes ----- Velocity domain feature collections are MeshNode-type features. These are produced from `lons` and `lats` pairs represented as multi-point geometries (projections onto the surface of the unit length sphere). These features are resolved into domain points and assigned plate IDs which are used to obtain the equivalent stage rotations of identified tectonic plates over a time interval (`delta_time`). Each velocity domain point and its stage rotation are used to calculate the point's plate velocity at a particular `time`. Obtained velocities for each domain point are represented in the north-east-down coordinate system. Parameters ---------- lons : array A 1D array of point data's longitudes. lats : array A 1D array of point data's latitudes. time : float The specific geological time (Ma) at which to calculate plate velocities. delta_time : float, default=1.0 The time increment used for generating partitioning plate stage rotations. 1.0Ma by default. Returns ------- all_velocities : 1D list of tuples For each velocity domain feature point, a tuple of (north, east, down) velocity components is generated and appended to a list of velocity data. The length of `all_velocities` is equivalent to the number of domain points resolved from the lat-lon array parameters. """ # Add points to a multipoint geometry time = float(time) multi_point = pygplates.MultiPointOnSphere( [(float(lat), float(lon)) for lat, lon in zip(lats, lons)] ) # Create a feature containing the multipoint feature, and defined as MeshNode type meshnode_feature = pygplates.Feature( pygplates.FeatureType.create_from_qualified_string("gpml:MeshNode") ) meshnode_feature.set_geometry(multi_point) meshnode_feature.set_name("Velocity Mesh Nodes from pygplates") velocity_domain_features = pygplates.FeatureCollection(meshnode_feature) # NB: at this point, the feature could be written to a file using # output_feature_collection.write('myfilename.gpmlz') # All domain points and associated (magnitude, azimuth, inclination) velocities for the current time. all_domain_points = [] all_velocities = [] # Partition our velocity domain features into our topological plate polygons at the current 'time'. plate_partitioner = pygplates.PlatePartitioner( self.topology_features, self.rotation_model, time ) for velocity_domain_feature in velocity_domain_features: # A velocity domain feature usually has a single geometry but we'll assume it can be any number. # Iterate over them all. for velocity_domain_geometry in velocity_domain_feature.get_geometries(): for velocity_domain_point in velocity_domain_geometry.get_points(): all_domain_points.append(velocity_domain_point) partitioning_plate = plate_partitioner.partition_point( velocity_domain_point ) if partitioning_plate: # We need the newly assigned plate ID # to get the equivalent stage rotation of that tectonic plate. partitioning_plate_id = ( partitioning_plate.get_feature().get_reconstruction_plate_id() ) # Get the stage rotation of partitioning plate from 'time + delta_time' to 'time'. equivalent_stage_rotation = self.rotation_model.get_rotation( time, partitioning_plate_id, time + delta_time ) # Calculate velocity at the velocity domain point. # This is from 'time + delta_time' to 'time' on the partitioning plate. velocity_vectors = pygplates.calculate_velocities( [velocity_domain_point], equivalent_stage_rotation, delta_time, ) # Convert global 3D velocity vectors to local (magnitude, azimuth, inclination) tuples # (one tuple per point). velocities = pygplates.LocalCartesian.convert_from_geocentric_to_north_east_down( [velocity_domain_point], velocity_vectors ) all_velocities.append( (velocities[0].get_x(), velocities[0].get_y()) ) else: all_velocities.append((0, 0)) return np.array(all_velocities) def create_motion_path( self, lons, lats, time_array, plate_id=None, anchor_plate_id=None, return_rate_of_motion=False, ): """Create a path of points to mark the trajectory of a plate's motion through geological time. Parameters ---------- lons : arr An array containing the longitudes of seed points on a plate in motion. lats : arr An array containing the latitudes of seed points on a plate in motion. time_array : arr An array of reconstruction times at which to determine the trajectory of a point on a plate. For example: import numpy as np min_time = 30 max_time = 100 time_step = 2.5 time_array = np.arange(min_time, max_time + time_step, time_step) plate_id : int, default=None The ID of the moving plate. If this is not passed, the plate ID of the seed points are ascertained using pygplates' `PlatePartitioner`. anchor_plate_id : int, default=0 The ID of the anchor plate. return_rate_of_motion : bool, default=False Choose whether to return the rate of plate motion through time for each Returns ------- rlons : ndarray An n-dimensional array with columns containing the longitudes of the seed points at each timestep in `time_array`. There are n columns for n seed points. rlats : ndarray An n-dimensional array with columns containing the latitudes of the seed points at each timestep in `time_array`. There are n columns for n seed points. StepTimes StepRates Examples -------- To access the latitudes and longitudes of each seed point's motion path: for i in np.arange(0,len(seed_points)): current_lons = lon[:,i] current_lats = lat[:,i] """ lons = np.atleast_1d(lons) lats = np.atleast_1d(lats) time_array = np.atleast_1d(time_array) # ndarrays to fill with reconstructed points and # rates of motion (if requested) rlons = np.empty((len(time_array), len(lons))) rlats = np.empty((len(time_array), len(lons))) if plate_id is None: query_plate_id = True else: query_plate_id = False plate_ids = np.ones(len(lons), dtype=int) * plate_id if anchor_plate_id is None: anchor_plate_id = self.anchor_plate_id seed_points = zip(lats, lons) if return_rate_of_motion is True: StepTimes = np.empty(((len(time_array) - 1) * 2, len(lons))) StepRates = np.empty(((len(time_array) - 1) * 2, len(lons))) for i, lat_lon in enumerate(seed_points): seed_points_at_digitisation_time = pygplates.PointOnSphere( pygplates.LatLonPoint(float(lat_lon[0]), float(lat_lon[1])) ) # Allocate the present-day plate ID to the PointOnSphere if # it was not given. if query_plate_id: plate_id = _tools.plate_partitioner_for_point( lat_lon, self.topology_features, self.rotation_model ) else: plate_id = plate_ids[i] # Create the motion path feature. enforce float and int for C++ signature. motion_path_feature = pygplates.Feature.create_motion_path( seed_points_at_digitisation_time, time_array, valid_time=(time_array.max(), time_array.min()), relative_plate=int(anchor_plate_id), reconstruction_plate_id=int(plate_id), ) reconstructed_motion_paths = self.reconstruct( motion_path_feature, to_time=0, reconstruct_type=pygplates.ReconstructType.motion_path, anchor_plate_id=int(anchor_plate_id), ) # Turn motion paths in to lat-lon coordinates for reconstructed_motion_path in reconstructed_motion_paths: trail = reconstructed_motion_path.get_motion_path().to_lat_lon_array() lon, lat = np.flipud(trail[:, 1]), np.flipud(trail[:, 0]) rlons[:, i] = lon rlats[:, i] = lat # Obtain step-plot coordinates for rate of motion if return_rate_of_motion is True: # Get timestep TimeStep = [] for j in range(len(time_array) - 1): diff = time_array[j + 1] - time_array[j] TimeStep.append(diff) # Iterate over each segment in the reconstructed motion path, get the distance travelled by the moving # plate relative to the fixed plate in each time step Dist = [] for reconstructed_motion_path in reconstructed_motion_paths: for ( segment ) in reconstructed_motion_path.get_motion_path().get_segments(): Dist.append( segment.get_arc_length() * _tools.geocentric_radius( segment.get_start_point().to_lat_lon()[0] ) / 1e3 ) # Note that the motion path coordinates come out starting with the oldest time and working forwards # So, to match our 'times' array, we flip the order Dist = np.flipud(Dist) # Get rate of motion as distance per Myr Rate = np.asarray(Dist) / TimeStep # Manipulate arrays to get a step plot StepRate = np.zeros(len(Rate) * 2) StepRate[::2] = Rate StepRate[1::2] = Rate StepTime = np.zeros(len(Rate) * 2) StepTime[::2] = time_array[:-1] StepTime[1::2] = time_array[1:] # Append the nth point's step time and step rate coordinates to the ndarray StepTimes[:, i] = StepTime StepRates[:, i] = StepRate * 0.1 # cm/yr # Obseleted by Lauren's changes above (though it is more efficient) # multiply arc length of the motion path segment by a latitude-dependent Earth radius # use latitude of the segment start point # distance.append( segment.get_arc_length() * _tools.geocentric_radius(segment.get_start_point().to_lat_lon()[0]) / 1e3) # rate = np.asarray(distance)/np.diff(time_array) # rates[:,i] = np.flipud(rate) # rates *= 0.1 # cm/yr if return_rate_of_motion is True: return ( np.squeeze(rlons), np.squeeze(rlats), np.squeeze(StepTimes), np.squeeze(StepRates), ) else: return np.squeeze(rlons), np.squeeze(rlats) def create_flowline( self, lons, lats, time_array, left_plate_ID, right_plate_ID, return_rate_of_motion=False, ): """Create a path of points to track plate motion away from spreading ridges over time using half-stage rotations. Parameters ---------- lons : arr An array of longitudes of points along spreading ridges. lats : arr An array of latitudes of points along spreading ridges. time_array : arr A list of times to obtain seed point locations at. left_plate_ID : int The plate ID of the polygon to the left of the spreading ridge. right_plate_ID : int The plate ID of the polygon to the right of the spreading ridge. return_rate_of_motion : bool, default False Choose whether to return a step time and step rate array for a step plot of motion. Returns ------- left_lon : ndarray The longitudes of the __left__ flowline for n seed points. There are n columns for n seed points, and m rows for m time steps in `time_array`. left_lat : ndarray The latitudes of the __left__ flowline of n seed points. There are n columns for n seed points, and m rows for m time steps in `time_array`. right_lon : ndarray The longitudes of the __right__ flowline of n seed points. There are n columns for n seed points, and m rows for m time steps in `time_array`. right_lat : ndarray The latitudes of the __right__ flowline of n seed points. There are n columns for n seed points, and m rows for m time steps in `time_array`. Examples -------- To access the ith seed point's left and right latitudes and longitudes: for i in np.arange(0,len(seed_points)): left_flowline_longitudes = left_lon[:,i] left_flowline_latitudes = left_lat[:,i] right_flowline_longitudes = right_lon[:,i] right_flowline_latitudes = right_lat[:,i] """ lats = np.atleast_1d(lats) lons = np.atleast_1d(lons) time_array = np.atleast_1d(time_array) seed_points = list(zip(lats, lons)) multi_point = pygplates.MultiPointOnSphere(seed_points) start = 0 if time_array[0] != 0: start = 1 time_array = np.hstack([[0], time_array]) # Create the flowline feature flowline_feature = pygplates.Feature.create_flowline( multi_point, time_array.tolist(), valid_time=(time_array.max(), time_array.min()), left_plate=left_plate_ID, right_plate=right_plate_ID, ) # reconstruct the flowline in present-day coordinates reconstructed_flowlines = self.reconstruct( flowline_feature, to_time=0, reconstruct_type=pygplates.ReconstructType.flowline, ) # Wrap things to the dateline, to avoid plotting artefacts. date_line_wrapper = pygplates.DateLineWrapper() # Create lat-lon ndarrays to store the left and right lats and lons of flowlines left_lon = np.empty((len(time_array), len(lons))) left_lat = np.empty((len(time_array), len(lons))) right_lon = np.empty((len(time_array), len(lons))) right_lat = np.empty((len(time_array), len(lons))) StepTimes = np.empty(((len(time_array) - 1) * 2, len(lons))) StepRates = np.empty(((len(time_array) - 1) * 2, len(lons))) # Iterate over the reconstructed flowlines. Each seed point results in a 'left' and 'right' flowline for i, reconstructed_flowline in enumerate(reconstructed_flowlines): # Get the points for the left flowline only left_latlon = reconstructed_flowline.get_left_flowline().to_lat_lon_array() left_lon[:, i] = left_latlon[:, 1] left_lat[:, i] = left_latlon[:, 0] # Repeat for the right flowline points right_latlon = ( reconstructed_flowline.get_right_flowline().to_lat_lon_array() ) right_lon[:, i] = right_latlon[:, 1] right_lat[:, i] = right_latlon[:, 0] if return_rate_of_motion: for i, reconstructed_motion_path in enumerate(reconstructed_flowlines): distance = [] for ( segment ) in reconstructed_motion_path.get_left_flowline().get_segments(): distance.append( segment.get_arc_length() * _tools.geocentric_radius( segment.get_start_point().to_lat_lon()[0] ) / 1e3 ) # Get rate of motion as distance per Myr # Need to multiply rate by 2, since flowlines give us half-spreading rate time_step = time_array[1] - time_array[0] Rate = ( np.asarray(distance) / time_step ) * 2 # since we created the flowline at X increment # Manipulate arrays to get a step plot StepRate = np.zeros(len(Rate) * 2) StepRate[::2] = Rate StepRate[1::2] = Rate StepTime = np.zeros(len(Rate) * 2) StepTime[::2] = time_array[:-1] StepTime[1::2] = time_array[1:] # Append the nth point's step time and step rate coordinates to the ndarray StepTimes[:, i] = StepTime StepRates[:, i] = StepRate * 0.1 # cm/yr return ( left_lon[start:], left_lat[start:], right_lon[start:], right_lat[start:], StepTimes, StepRates, ) else: return ( left_lon[start:], left_lat[start:], right_lon[start:], right_lat[start:], )Methods
def create_flowline(self, lons, lats, time_array, left_plate_ID, right_plate_ID, return_rate_of_motion=False)-
Create a path of points to track plate motion away from spreading ridges over time using half-stage rotations.
Parameters
lons:arr- An array of longitudes of points along spreading ridges.
lats:arr- An array of latitudes of points along spreading ridges.
time_array:arr- A list of times to obtain seed point locations at.
left_plate_ID:int- The plate ID of the polygon to the left of the spreading ridge.
right_plate_ID:int- The plate ID of the polygon to the right of the spreading ridge.
return_rate_of_motion:bool, defaultFalse- Choose whether to return a step time and step rate array for a step plot of motion.
Returns
left_lon:ndarray- The longitudes of the left flowline for n seed points.
There are n columns for n seed points, and m rows
for m time steps in
time_array. left_lat:ndarray- The latitudes of the left flowline of n seed points.
There are n columns for n seed points, and m rows
for m time steps in
time_array. right_lon:ndarray- The longitudes of the right flowline of n seed points.
There are n columns for n seed points, and m rows
for m time steps in
time_array. right_lat:ndarray- The latitudes of the right flowline of n seed points.
There are n columns for n seed points, and m rows
for m time steps in
time_array.
Examples
To access the ith seed point's left and right latitudes and longitudes:
for i in np.arange(0,len(seed_points)): left_flowline_longitudes = left_lon[:,i] left_flowline_latitudes = left_lat[:,i] right_flowline_longitudes = right_lon[:,i] right_flowline_latitudes = right_lat[:,i]Expand source code
def create_flowline( self, lons, lats, time_array, left_plate_ID, right_plate_ID, return_rate_of_motion=False, ): """Create a path of points to track plate motion away from spreading ridges over time using half-stage rotations. Parameters ---------- lons : arr An array of longitudes of points along spreading ridges. lats : arr An array of latitudes of points along spreading ridges. time_array : arr A list of times to obtain seed point locations at. left_plate_ID : int The plate ID of the polygon to the left of the spreading ridge. right_plate_ID : int The plate ID of the polygon to the right of the spreading ridge. return_rate_of_motion : bool, default False Choose whether to return a step time and step rate array for a step plot of motion. Returns ------- left_lon : ndarray The longitudes of the __left__ flowline for n seed points. There are n columns for n seed points, and m rows for m time steps in `time_array`. left_lat : ndarray The latitudes of the __left__ flowline of n seed points. There are n columns for n seed points, and m rows for m time steps in `time_array`. right_lon : ndarray The longitudes of the __right__ flowline of n seed points. There are n columns for n seed points, and m rows for m time steps in `time_array`. right_lat : ndarray The latitudes of the __right__ flowline of n seed points. There are n columns for n seed points, and m rows for m time steps in `time_array`. Examples -------- To access the ith seed point's left and right latitudes and longitudes: for i in np.arange(0,len(seed_points)): left_flowline_longitudes = left_lon[:,i] left_flowline_latitudes = left_lat[:,i] right_flowline_longitudes = right_lon[:,i] right_flowline_latitudes = right_lat[:,i] """ lats = np.atleast_1d(lats) lons = np.atleast_1d(lons) time_array = np.atleast_1d(time_array) seed_points = list(zip(lats, lons)) multi_point = pygplates.MultiPointOnSphere(seed_points) start = 0 if time_array[0] != 0: start = 1 time_array = np.hstack([[0], time_array]) # Create the flowline feature flowline_feature = pygplates.Feature.create_flowline( multi_point, time_array.tolist(), valid_time=(time_array.max(), time_array.min()), left_plate=left_plate_ID, right_plate=right_plate_ID, ) # reconstruct the flowline in present-day coordinates reconstructed_flowlines = self.reconstruct( flowline_feature, to_time=0, reconstruct_type=pygplates.ReconstructType.flowline, ) # Wrap things to the dateline, to avoid plotting artefacts. date_line_wrapper = pygplates.DateLineWrapper() # Create lat-lon ndarrays to store the left and right lats and lons of flowlines left_lon = np.empty((len(time_array), len(lons))) left_lat = np.empty((len(time_array), len(lons))) right_lon = np.empty((len(time_array), len(lons))) right_lat = np.empty((len(time_array), len(lons))) StepTimes = np.empty(((len(time_array) - 1) * 2, len(lons))) StepRates = np.empty(((len(time_array) - 1) * 2, len(lons))) # Iterate over the reconstructed flowlines. Each seed point results in a 'left' and 'right' flowline for i, reconstructed_flowline in enumerate(reconstructed_flowlines): # Get the points for the left flowline only left_latlon = reconstructed_flowline.get_left_flowline().to_lat_lon_array() left_lon[:, i] = left_latlon[:, 1] left_lat[:, i] = left_latlon[:, 0] # Repeat for the right flowline points right_latlon = ( reconstructed_flowline.get_right_flowline().to_lat_lon_array() ) right_lon[:, i] = right_latlon[:, 1] right_lat[:, i] = right_latlon[:, 0] if return_rate_of_motion: for i, reconstructed_motion_path in enumerate(reconstructed_flowlines): distance = [] for ( segment ) in reconstructed_motion_path.get_left_flowline().get_segments(): distance.append( segment.get_arc_length() * _tools.geocentric_radius( segment.get_start_point().to_lat_lon()[0] ) / 1e3 ) # Get rate of motion as distance per Myr # Need to multiply rate by 2, since flowlines give us half-spreading rate time_step = time_array[1] - time_array[0] Rate = ( np.asarray(distance) / time_step ) * 2 # since we created the flowline at X increment # Manipulate arrays to get a step plot StepRate = np.zeros(len(Rate) * 2) StepRate[::2] = Rate StepRate[1::2] = Rate StepTime = np.zeros(len(Rate) * 2) StepTime[::2] = time_array[:-1] StepTime[1::2] = time_array[1:] # Append the nth point's step time and step rate coordinates to the ndarray StepTimes[:, i] = StepTime StepRates[:, i] = StepRate * 0.1 # cm/yr return ( left_lon[start:], left_lat[start:], right_lon[start:], right_lat[start:], StepTimes, StepRates, ) else: return ( left_lon[start:], left_lat[start:], right_lon[start:], right_lat[start:], ) def create_motion_path(self, lons, lats, time_array, plate_id=None, anchor_plate_id=None, return_rate_of_motion=False)-
Create a path of points to mark the trajectory of a plate's motion through geological time.
Parameters
lons:arr- An array containing the longitudes of seed points on a plate in motion.
lats:arr- An array containing the latitudes of seed points on a plate in motion.
time_array:arr- An array of reconstruction times at which to determine the trajectory
of a point on a plate. For example:
import numpy as np min_time = 30 max_time = 100 time_step = 2.5 time_array = np.arange(min_time, max_time + time_step, time_step) plate_id:int, default=None- The ID of the moving plate. If this is not passed, the plate ID of the
seed points are ascertained using pygplates'
PlatePartitioner. anchor_plate_id:int, default=0- The ID of the anchor plate.
return_rate_of_motion:bool, default=False- Choose whether to return the rate of plate motion through time for each
Returns
rlons:ndarray- An n-dimensional array with columns containing the longitudes of
the seed points at each timestep in
time_array. There are n columns for n seed points. rlats:ndarray- An n-dimensional array with columns containing the latitudes of
the seed points at each timestep in
time_array. There are n columns for n seed points. StepTimesStepRates
Examples
To access the latitudes and longitudes of each seed point's motion path:
for i in np.arange(0,len(seed_points)): current_lons = lon[:,i] current_lats = lat[:,i]Expand source code
def create_motion_path( self, lons, lats, time_array, plate_id=None, anchor_plate_id=None, return_rate_of_motion=False, ): """Create a path of points to mark the trajectory of a plate's motion through geological time. Parameters ---------- lons : arr An array containing the longitudes of seed points on a plate in motion. lats : arr An array containing the latitudes of seed points on a plate in motion. time_array : arr An array of reconstruction times at which to determine the trajectory of a point on a plate. For example: import numpy as np min_time = 30 max_time = 100 time_step = 2.5 time_array = np.arange(min_time, max_time + time_step, time_step) plate_id : int, default=None The ID of the moving plate. If this is not passed, the plate ID of the seed points are ascertained using pygplates' `PlatePartitioner`. anchor_plate_id : int, default=0 The ID of the anchor plate. return_rate_of_motion : bool, default=False Choose whether to return the rate of plate motion through time for each Returns ------- rlons : ndarray An n-dimensional array with columns containing the longitudes of the seed points at each timestep in `time_array`. There are n columns for n seed points. rlats : ndarray An n-dimensional array with columns containing the latitudes of the seed points at each timestep in `time_array`. There are n columns for n seed points. StepTimes StepRates Examples -------- To access the latitudes and longitudes of each seed point's motion path: for i in np.arange(0,len(seed_points)): current_lons = lon[:,i] current_lats = lat[:,i] """ lons = np.atleast_1d(lons) lats = np.atleast_1d(lats) time_array = np.atleast_1d(time_array) # ndarrays to fill with reconstructed points and # rates of motion (if requested) rlons = np.empty((len(time_array), len(lons))) rlats = np.empty((len(time_array), len(lons))) if plate_id is None: query_plate_id = True else: query_plate_id = False plate_ids = np.ones(len(lons), dtype=int) * plate_id if anchor_plate_id is None: anchor_plate_id = self.anchor_plate_id seed_points = zip(lats, lons) if return_rate_of_motion is True: StepTimes = np.empty(((len(time_array) - 1) * 2, len(lons))) StepRates = np.empty(((len(time_array) - 1) * 2, len(lons))) for i, lat_lon in enumerate(seed_points): seed_points_at_digitisation_time = pygplates.PointOnSphere( pygplates.LatLonPoint(float(lat_lon[0]), float(lat_lon[1])) ) # Allocate the present-day plate ID to the PointOnSphere if # it was not given. if query_plate_id: plate_id = _tools.plate_partitioner_for_point( lat_lon, self.topology_features, self.rotation_model ) else: plate_id = plate_ids[i] # Create the motion path feature. enforce float and int for C++ signature. motion_path_feature = pygplates.Feature.create_motion_path( seed_points_at_digitisation_time, time_array, valid_time=(time_array.max(), time_array.min()), relative_plate=int(anchor_plate_id), reconstruction_plate_id=int(plate_id), ) reconstructed_motion_paths = self.reconstruct( motion_path_feature, to_time=0, reconstruct_type=pygplates.ReconstructType.motion_path, anchor_plate_id=int(anchor_plate_id), ) # Turn motion paths in to lat-lon coordinates for reconstructed_motion_path in reconstructed_motion_paths: trail = reconstructed_motion_path.get_motion_path().to_lat_lon_array() lon, lat = np.flipud(trail[:, 1]), np.flipud(trail[:, 0]) rlons[:, i] = lon rlats[:, i] = lat # Obtain step-plot coordinates for rate of motion if return_rate_of_motion is True: # Get timestep TimeStep = [] for j in range(len(time_array) - 1): diff = time_array[j + 1] - time_array[j] TimeStep.append(diff) # Iterate over each segment in the reconstructed motion path, get the distance travelled by the moving # plate relative to the fixed plate in each time step Dist = [] for reconstructed_motion_path in reconstructed_motion_paths: for ( segment ) in reconstructed_motion_path.get_motion_path().get_segments(): Dist.append( segment.get_arc_length() * _tools.geocentric_radius( segment.get_start_point().to_lat_lon()[0] ) / 1e3 ) # Note that the motion path coordinates come out starting with the oldest time and working forwards # So, to match our 'times' array, we flip the order Dist = np.flipud(Dist) # Get rate of motion as distance per Myr Rate = np.asarray(Dist) / TimeStep # Manipulate arrays to get a step plot StepRate = np.zeros(len(Rate) * 2) StepRate[::2] = Rate StepRate[1::2] = Rate StepTime = np.zeros(len(Rate) * 2) StepTime[::2] = time_array[:-1] StepTime[1::2] = time_array[1:] # Append the nth point's step time and step rate coordinates to the ndarray StepTimes[:, i] = StepTime StepRates[:, i] = StepRate * 0.1 # cm/yr # Obseleted by Lauren's changes above (though it is more efficient) # multiply arc length of the motion path segment by a latitude-dependent Earth radius # use latitude of the segment start point # distance.append( segment.get_arc_length() * _tools.geocentric_radius(segment.get_start_point().to_lat_lon()[0]) / 1e3) # rate = np.asarray(distance)/np.diff(time_array) # rates[:,i] = np.flipud(rate) # rates *= 0.1 # cm/yr if return_rate_of_motion is True: return ( np.squeeze(rlons), np.squeeze(rlats), np.squeeze(StepTimes), np.squeeze(StepRates), ) else: return np.squeeze(rlons), np.squeeze(rlats) def get_point_velocities(self, lons, lats, time, delta_time=1.0)-
Generates a velocity domain feature collection, resolves them into points, and calculates the north and east components of the velocity vector for each point in the domain at a particular geological
time.Notes
Velocity domain feature collections are MeshNode-type features. These are produced from
lonsandlatspairs represented as multi-point geometries (projections onto the surface of the unit length sphere). These features are resolved into domain points and assigned plate IDs which are used to obtain the equivalent stage rotations of identified tectonic plates over a time interval (delta_time). Each velocity domain point and its stage rotation are used to calculate the point's plate velocity at a particulartime. Obtained velocities for each domain point are represented in the north-east-down coordinate system.Parameters
lons:array- A 1D array of point data's longitudes.
lats:array- A 1D array of point data's latitudes.
time:float- The specific geological time (Ma) at which to calculate plate velocities.
delta_time:float, default=1.0- The time increment used for generating partitioning plate stage rotations. 1.0Ma by default.
Returns
all_velocities:1D listoftuples- For each velocity domain feature point, a tuple of (north, east, down) velocity components is generated and
appended to a list of velocity data. The length of
all_velocitiesis equivalent to the number of domain points resolved from the lat-lon array parameters.
Expand source code
def get_point_velocities(self, lons, lats, time, delta_time=1.0): """Generates a velocity domain feature collection, resolves them into points, and calculates the north and east components of the velocity vector for each point in the domain at a particular geological `time`. Notes ----- Velocity domain feature collections are MeshNode-type features. These are produced from `lons` and `lats` pairs represented as multi-point geometries (projections onto the surface of the unit length sphere). These features are resolved into domain points and assigned plate IDs which are used to obtain the equivalent stage rotations of identified tectonic plates over a time interval (`delta_time`). Each velocity domain point and its stage rotation are used to calculate the point's plate velocity at a particular `time`. Obtained velocities for each domain point are represented in the north-east-down coordinate system. Parameters ---------- lons : array A 1D array of point data's longitudes. lats : array A 1D array of point data's latitudes. time : float The specific geological time (Ma) at which to calculate plate velocities. delta_time : float, default=1.0 The time increment used for generating partitioning plate stage rotations. 1.0Ma by default. Returns ------- all_velocities : 1D list of tuples For each velocity domain feature point, a tuple of (north, east, down) velocity components is generated and appended to a list of velocity data. The length of `all_velocities` is equivalent to the number of domain points resolved from the lat-lon array parameters. """ # Add points to a multipoint geometry time = float(time) multi_point = pygplates.MultiPointOnSphere( [(float(lat), float(lon)) for lat, lon in zip(lats, lons)] ) # Create a feature containing the multipoint feature, and defined as MeshNode type meshnode_feature = pygplates.Feature( pygplates.FeatureType.create_from_qualified_string("gpml:MeshNode") ) meshnode_feature.set_geometry(multi_point) meshnode_feature.set_name("Velocity Mesh Nodes from pygplates") velocity_domain_features = pygplates.FeatureCollection(meshnode_feature) # NB: at this point, the feature could be written to a file using # output_feature_collection.write('myfilename.gpmlz') # All domain points and associated (magnitude, azimuth, inclination) velocities for the current time. all_domain_points = [] all_velocities = [] # Partition our velocity domain features into our topological plate polygons at the current 'time'. plate_partitioner = pygplates.PlatePartitioner( self.topology_features, self.rotation_model, time ) for velocity_domain_feature in velocity_domain_features: # A velocity domain feature usually has a single geometry but we'll assume it can be any number. # Iterate over them all. for velocity_domain_geometry in velocity_domain_feature.get_geometries(): for velocity_domain_point in velocity_domain_geometry.get_points(): all_domain_points.append(velocity_domain_point) partitioning_plate = plate_partitioner.partition_point( velocity_domain_point ) if partitioning_plate: # We need the newly assigned plate ID # to get the equivalent stage rotation of that tectonic plate. partitioning_plate_id = ( partitioning_plate.get_feature().get_reconstruction_plate_id() ) # Get the stage rotation of partitioning plate from 'time + delta_time' to 'time'. equivalent_stage_rotation = self.rotation_model.get_rotation( time, partitioning_plate_id, time + delta_time ) # Calculate velocity at the velocity domain point. # This is from 'time + delta_time' to 'time' on the partitioning plate. velocity_vectors = pygplates.calculate_velocities( [velocity_domain_point], equivalent_stage_rotation, delta_time, ) # Convert global 3D velocity vectors to local (magnitude, azimuth, inclination) tuples # (one tuple per point). velocities = pygplates.LocalCartesian.convert_from_geocentric_to_north_east_down( [velocity_domain_point], velocity_vectors ) all_velocities.append( (velocities[0].get_x(), velocities[0].get_y()) ) else: all_velocities.append((0, 0)) return np.array(all_velocities) def reconstruct(self, feature, to_time, from_time=0, anchor_plate_id=None, **kwargs)-
Reconstructs regular geological features, motion paths or flowlines to a specific geological time.
Parameters
feature:str,orinstanceof<pygplates.FeatureCollection>,or<pygplates.Feature>,orsequenceof<pygplates.Feature>- The geological features to reconstruct. Can be provided as a feature collection, or filename, or feature, or sequence of features, or a sequence (eg, a list or tuple) of any combination of those four types.
to_time:float,or:class:GeoTimeInstant``- The specific geological time to reconstruct to.
from_time:float, default=0- The specific geological time to reconstruct from. By default, this is set to present day. Raises
NotImplementedErroriffrom_timeis not set to 0.0 Ma (present day). anchor_plate_id:int, default=None- Reconstruct features with respect to a certain anchor plate. By default, reconstructions are made with respect to the absolute reference frame (anchor_plate_id = 0), like a stationary object in the mantle, unless otherwise specified.
**reconstruct_type:ReconstructType, default=ReconstructType.feature_geometry- The specific reconstruction type to generate based on input feature geometry type. Can be provided as
pygplates.ReconstructType.feature_geometry to only reconstruct regular feature geometries, or
pygplates.ReconstructType.motion_path to only reconstruct motion path features, or
pygplates.ReconstructType.flowline to only reconstruct flowline features.
Generates :class:
reconstructed feature geometries<ReconstructedFeatureGeometry>’, or :class:reconstructed motion paths’, or :class:`reconstructed flowlines ’ respectively. **group_with_feature:bool, default=False- Used to group reconstructed geometries with their features. This can be useful when a feature has more than one
geometry and hence more than one reconstructed geometry. The output reconstructed_geometries then becomes a
list of tuples where each tuple contains a :class:
feature<Feature>and alistof reconstructed geometries. Note: this keyword argument only applies when reconstructed_geometries is a list because exported files are always grouped with their features. This is applicable to all ReconstructType features. **export_wrap_to_dateline:bool, default=True- Wrap/clip reconstructed geometries to the dateline.
Returns
reconstructed_features:list- Reconstructed geological features (generated by the reconstruction) are appended to the list.
The reconstructed geometries are output in the same order as that of their respective input features (in the
parameter “features”). The order across input feature collections is also retained. This happens regardless
of whether features and reconstructed_features include files or not. Note: if keyword argument
group_with_feature=True then the list contains tuples that group each :class:
feature<Feature>with a list of its reconstructed geometries.
Raises
NotImplementedError- if the starting time for reconstruction
from_timeis not equal to 0.0.
Expand source code
def reconstruct( self, feature, to_time, from_time=0, anchor_plate_id=None, **kwargs ): """Reconstructs regular geological features, motion paths or flowlines to a specific geological time. Parameters ---------- feature : str, or instance of <pygplates.FeatureCollection>, or <pygplates.Feature>, or sequence of <pygplates.Feature> The geological features to reconstruct. Can be provided as a feature collection, or filename, or feature, or sequence of features, or a sequence (eg, a list or tuple) of any combination of those four types. to_time : float, or :class:`GeoTimeInstant` The specific geological time to reconstruct to. from_time : float, default=0 The specific geological time to reconstruct from. By default, this is set to present day. Raises `NotImplementedError` if `from_time` is not set to 0.0 Ma (present day). anchor_plate_id : int, default=None Reconstruct features with respect to a certain anchor plate. By default, reconstructions are made with respect to the absolute reference frame (anchor_plate_id = 0), like a stationary object in the mantle, unless otherwise specified. **reconstruct_type : ReconstructType, default=ReconstructType.feature_geometry The specific reconstruction type to generate based on input feature geometry type. Can be provided as pygplates.ReconstructType.feature_geometry to only reconstruct regular feature geometries, or pygplates.ReconstructType.motion_path to only reconstruct motion path features, or pygplates.ReconstructType.flowline to only reconstruct flowline features. Generates :class:`reconstructed feature geometries<ReconstructedFeatureGeometry>’, or :class:`reconstructed motion paths<ReconstructedMotionPath>’, or :class:`reconstructed flowlines<ReconstructedFlowline>’ respectively. **group_with_feature : bool, default=False Used to group reconstructed geometries with their features. This can be useful when a feature has more than one geometry and hence more than one reconstructed geometry. The output *reconstructed_geometries* then becomes a list of tuples where each tuple contains a :class:`feature<Feature>` and a ``list`` of reconstructed geometries. Note: this keyword argument only applies when *reconstructed_geometries* is a list because exported files are always grouped with their features. This is applicable to all ReconstructType features. **export_wrap_to_dateline : bool, default=True Wrap/clip reconstructed geometries to the dateline. Returns ------- reconstructed_features : list Reconstructed geological features (generated by the reconstruction) are appended to the list. The reconstructed geometries are output in the same order as that of their respective input features (in the parameter “features”). The order across input feature collections is also retained. This happens regardless of whether *features* and *reconstructed_features* include files or not. Note: if keyword argument group_with_feature=True then the list contains tuples that group each :class:`feature<Feature>` with a list of its reconstructed geometries. Raises ------ NotImplementedError if the starting time for reconstruction `from_time` is not equal to 0.0. """ from_time, to_time = float(from_time), float(to_time) reconstructed_features = [] if not anchor_plate_id: anchor_plate_id = self.anchor_plate_id pygplates.reconstruct( feature, self.rotation_model, reconstructed_features, to_time, anchor_plate_id=anchor_plate_id, **kwargs ) return reconstructed_features def tessellate_mid_ocean_ridges(self, time, tessellation_threshold_radians=0.001, ignore_warnings=False, return_geodataframe=False, **kwargs)-
Samples points along resolved spreading features (e.g. mid-ocean ridges) and calculates spreading rates and lengths of ridge segments at a particular geological time.
Resolves topologies at
time, tessellates all resolved spreading features to within 'tessellation_threshold_radians' radians. Returns a 4-column vertically stacked tuple with the following data.- Col. 0 - longitude of sampled ridge point
- Col. 1 - latitude of sampled ridge point
- Col. 2 - spreading velocity magnitude (in cm/yr)
- Col. 3 - length of arc segment (in degrees) that current point is on
All spreading feature types are considered. The transform segments of spreading features are ignored. Note: by default, the function assumes that a segment can deviate 45 degrees from the stage pole before it is considered a transform segment.
Parameters
time:float- The reconstruction time (Ma) at which to query subduction convergence.
tessellation_threshold_radians:float, default=0.001- The threshold sampling distance along the subducting trench (in radians).
ignore_warnings:bool, default=False- Choose to ignore warnings from Plate Tectonic Tools' ridge_spreading_rate workflow.
Returns
ridge_data:a listofvertically-stacked tuples-
The results for all tessellated points sampled along the trench. The size of the returned list is equal to the number of tessellated points. Each tuple in the list corresponds to a tessellated point and has the following tuple items:
- longitude of sampled point
- latitude of sampled point
- spreading velocity magnitude (in cm/yr)
- length of arc segment (in degrees) that current point is on
Expand source code
def tessellate_mid_ocean_ridges( self, time, tessellation_threshold_radians=0.001, ignore_warnings=False, return_geodataframe=False, **kwargs ): """Samples points along resolved spreading features (e.g. mid-ocean ridges) and calculates spreading rates and lengths of ridge segments at a particular geological time. Resolves topologies at `time`, tessellates all resolved spreading features to within 'tessellation_threshold_radians' radians. Returns a 4-column vertically stacked tuple with the following data. * Col. 0 - longitude of sampled ridge point * Col. 1 - latitude of sampled ridge point * Col. 2 - spreading velocity magnitude (in cm/yr) * Col. 3 - length of arc segment (in degrees) that current point is on All spreading feature types are considered. The transform segments of spreading features are ignored. Note: by default, the function assumes that a segment can deviate 45 degrees from the stage pole before it is considered a transform segment. Parameters ---------- time : float The reconstruction time (Ma) at which to query subduction convergence. tessellation_threshold_radians : float, default=0.001 The threshold sampling distance along the subducting trench (in radians). ignore_warnings : bool, default=False Choose to ignore warnings from Plate Tectonic Tools' ridge_spreading_rate workflow. Returns ------- ridge_data : a list of vertically-stacked tuples The results for all tessellated points sampled along the trench. The size of the returned list is equal to the number of tessellated points. Each tuple in the list corresponds to a tessellated point and has the following tuple items: * longitude of sampled point * latitude of sampled point * spreading velocity magnitude (in cm/yr) * length of arc segment (in degrees) that current point is on """ from . import ptt as _ptt anchor_plate_id = kwargs.pop("anchor_plate_id", self.anchor_plate_id) if ignore_warnings: with warnings.catch_warnings(): warnings.simplefilter("ignore") spreading_feature_types = [pygplates.FeatureType.gpml_mid_ocean_ridge] ridge_data = _ptt.ridge_spreading_rate.spreading_rates( self.rotation_model, self.topology_features, float(time), tessellation_threshold_radians, spreading_feature_types, anchor_plate_id=anchor_plate_id, **kwargs ) else: spreading_feature_types = [pygplates.FeatureType.gpml_mid_ocean_ridge] ridge_data = _ptt.ridge_spreading_rate.spreading_rates( self.rotation_model, self.topology_features, float(time), tessellation_threshold_radians, spreading_feature_types, anchor_plate_id=anchor_plate_id, **kwargs ) if not ridge_data: # the _ptt.ridge_spreading_rate.spreading_rates might return None return ridge_data = np.vstack(ridge_data) if return_geodataframe: import geopandas as gpd from shapely import geometry points = [ geometry.Point(lon, lat) for lon, lat in zip(ridge_data[:, 0], ridge_data[:, 1]) ] gdf = gpd.GeoDataFrame( { "geometry": points, "velocity (cm/yr)": ridge_data[:, 2], "length (degrees)": ridge_data[:, 3], }, geometry="geometry", ) return gdf else: return ridge_data def tessellate_subduction_zones(self, time, tessellation_threshold_radians=0.001, ignore_warnings=False, return_geodataframe=False, **kwargs)-
Samples points along subduction zone trenches and obtains subduction data at a particular geological time.
Resolves topologies at
time, tessellates all resolved subducting features to within 'tessellation_threshold_radians' radians and obtains the following information for each sampled point along a trench:tessellate_subduction_zonesreturns a list of 10 vertically-stacked tuples with the following data per sampled trench point:- Col. 0 - longitude of sampled trench point
- Col. 1 - latitude of sampled trench point
- Col. 2 - subducting convergence (relative to trench) velocity magnitude (in cm/yr)
- Col. 3 - subducting convergence velocity obliquity angle (angle between trench normal vector and convergence velocity vector)
- Col. 4 - trench absolute (relative to anchor plate) velocity magnitude (in cm/yr)
- Col. 5 - trench absolute velocity obliquity angle (angle between trench normal vector and trench absolute velocity vector)
- Col. 6 - length of arc segment (in degrees) that current point is on
- Col. 7 - trench normal azimuth angle (clockwise starting at North, ie, 0 to 360 degrees) at current point
- Col. 8 - subducting plate ID
- Col. 9 - trench plate ID
Parameters
time:float- The reconstruction time (Ma) at which to query subduction convergence.
tessellation_threshold_radians:float, default=0.001- The threshold sampling distance along the subducting trench (in radians).
Returns
subduction_data:a listofvertically-stacked tuples-
The results for all tessellated points sampled along the trench. The size of the returned list is equal to the number of tessellated points. Each tuple in the list corresponds to a tessellated point and has the following tuple items:
- Col. 0 - longitude of sampled trench point
- Col. 1 - latitude of sampled trench point
- Col. 2 - subducting convergence (relative to trench) velocity magnitude (in cm/yr)
- Col. 3 - subducting convergence velocity obliquity angle (angle between trench normal vector and convergence velocity vector)
- Col. 4 - trench absolute (relative to anchor plate) velocity magnitude (in cm/yr)
- Col. 5 - trench absolute velocity obliquity angle (angle between trench normal vector and trench absolute velocity vector)
- Col. 6 - length of arc segment (in degrees) that current point is on
- Col. 7 - trench normal azimuth angle (clockwise starting at North, ie, 0 to 360 degrees) at current point
- Col. 8 - subducting plate ID
- Col. 9 - trench plate ID
Notes
Each sampled point in the output is the midpoint of a great circle arc between two adjacent points in the trench polyline. The trench normal vector used in the obliquity calculations is perpendicular to the great circle arc of each point (arc midpoint) and pointing towards the overriding plate (rather than away from it).
Each trench is sampled at approximately uniform intervals along its length (specified via a threshold sampling distance). The sampling along the entire length of a trench is not exactly uniform. Each segment along a trench is sampled such that the samples have a uniform spacing that is less than or equal to the threshold sampling distance. However each segment in a trench might have a slightly different spacing distance (since segment lengths are not integer multiples of the threshold sampling distance).
The trench normal (at each arc segment mid-point) always points towards the overriding plate. The obliquity angles are in the range (-180 180). The range (0, 180) goes clockwise (when viewed from above the Earth) from the trench normal direction to the velocity vector. The range (0, -180) goes counter-clockwise. You can change the range (-180, 180) to the range (0, 360) by adding 360 to negative angles. The trench normal is perpendicular to the trench and pointing toward the overriding plate.
Note that the convergence velocity magnitude is negative if the plates are diverging (if convergence obliquity angle is greater than 90 or less than -90). And note that the absolute velocity magnitude is negative if the trench (subduction zone) is moving towards the overriding plate (if absolute obliquity angle is less than 90 or greater than -90) - note that this ignores the kinematics of the subducting plate.
The delta time interval used for velocity calculations is, by default, assumed to be 1Ma.
Expand source code
def tessellate_subduction_zones( self, time, tessellation_threshold_radians=0.001, ignore_warnings=False, return_geodataframe=False, **kwargs ): """Samples points along subduction zone trenches and obtains subduction data at a particular geological time. Resolves topologies at `time`, tessellates all resolved subducting features to within 'tessellation_threshold_radians' radians and obtains the following information for each sampled point along a trench: `tessellate_subduction_zones` returns a list of 10 vertically-stacked tuples with the following data per sampled trench point: * Col. 0 - longitude of sampled trench point * Col. 1 - latitude of sampled trench point * Col. 2 - subducting convergence (relative to trench) velocity magnitude (in cm/yr) * Col. 3 - subducting convergence velocity obliquity angle (angle between trench normal vector and convergence velocity vector) * Col. 4 - trench absolute (relative to anchor plate) velocity magnitude (in cm/yr) * Col. 5 - trench absolute velocity obliquity angle (angle between trench normal vector and trench absolute velocity vector) * Col. 6 - length of arc segment (in degrees) that current point is on * Col. 7 - trench normal azimuth angle (clockwise starting at North, ie, 0 to 360 degrees) at current point * Col. 8 - subducting plate ID * Col. 9 - trench plate ID Parameters ---------- time : float The reconstruction time (Ma) at which to query subduction convergence. tessellation_threshold_radians : float, default=0.001 The threshold sampling distance along the subducting trench (in radians). Returns ------- subduction_data : a list of vertically-stacked tuples The results for all tessellated points sampled along the trench. The size of the returned list is equal to the number of tessellated points. Each tuple in the list corresponds to a tessellated point and has the following tuple items: * Col. 0 - longitude of sampled trench point * Col. 1 - latitude of sampled trench point * Col. 2 - subducting convergence (relative to trench) velocity magnitude (in cm/yr) * Col. 3 - subducting convergence velocity obliquity angle (angle between trench normal vector and convergence velocity vector) * Col. 4 - trench absolute (relative to anchor plate) velocity magnitude (in cm/yr) * Col. 5 - trench absolute velocity obliquity angle (angle between trench normal vector and trench absolute velocity vector) * Col. 6 - length of arc segment (in degrees) that current point is on * Col. 7 - trench normal azimuth angle (clockwise starting at North, ie, 0 to 360 degrees) at current point * Col. 8 - subducting plate ID * Col. 9 - trench plate ID Notes ----- Each sampled point in the output is the midpoint of a great circle arc between two adjacent points in the trench polyline. The trench normal vector used in the obliquity calculations is perpendicular to the great circle arc of each point (arc midpoint) and pointing towards the overriding plate (rather than away from it). Each trench is sampled at approximately uniform intervals along its length (specified via a threshold sampling distance). The sampling along the entire length of a trench is not exactly uniform. Each segment along a trench is sampled such that the samples have a uniform spacing that is less than or equal to the threshold sampling distance. However each segment in a trench might have a slightly different spacing distance (since segment lengths are not integer multiples of the threshold sampling distance). The trench normal (at each arc segment mid-point) always points *towards* the overriding plate. The obliquity angles are in the range (-180 180). The range (0, 180) goes clockwise (when viewed from above the Earth) from the trench normal direction to the velocity vector. The range (0, -180) goes counter-clockwise. You can change the range (-180, 180) to the range (0, 360) by adding 360 to negative angles. The trench normal is perpendicular to the trench and pointing toward the overriding plate. Note that the convergence velocity magnitude is negative if the plates are diverging (if convergence obliquity angle is greater than 90 or less than -90). And note that the absolute velocity magnitude is negative if the trench (subduction zone) is moving towards the overriding plate (if absolute obliquity angle is less than 90 or greater than -90) - note that this ignores the kinematics of the subducting plate. The delta time interval used for velocity calculations is, by default, assumed to be 1Ma. """ from . import ptt as _ptt anchor_plate_id = kwargs.pop("anchor_plate_id", self.anchor_plate_id) if ignore_warnings: with warnings.catch_warnings(): warnings.simplefilter("ignore") subduction_data = _ptt.subduction_convergence.subduction_convergence( self.rotation_model, self.topology_features, tessellation_threshold_radians, float(time), anchor_plate_id=anchor_plate_id, **kwargs ) else: subduction_data = _ptt.subduction_convergence.subduction_convergence( self.rotation_model, self.topology_features, tessellation_threshold_radians, float(time), anchor_plate_id=anchor_plate_id, **kwargs ) subduction_data = np.vstack(subduction_data) if return_geodataframe: import geopandas as gpd from shapely import geometry coords = [ geometry.Point(lon, lat) for lon, lat in zip(subduction_data[:, 0], subduction_data[:, 1]) ] d = {"geometry": coords} labels = [ "convergence velocity (cm/yr)", "convergence obliquity angle (degrees)", "trench velocity (cm/yr)", "trench obliquity angle (degrees)", "length (degrees)", "trench normal angle (degrees)", "subducting plate ID", "overriding plate ID", ] for i, label in enumerate(labels): index = 2 + i d[label] = subduction_data[:, index] gdf = gpd.GeoDataFrame(d, geometry="geometry") return gdf else: return subduction_data def total_continental_arc_length(self, time, continental_grid, trench_arc_distance, ignore_warnings=True)-
Calculates the total length of all global continental arcs (km) at the specified geological time (Ma).
Uses Plate Tectonic Tools'
subduction_convergenceworkflow to sample a given plate model's trench features into point features and obtain their subduction polarities. The resolved points are projected out by thetrench_arc_distanceand their new locations are linearly interpolated onto the suppliedcontinental_grid. If the projected trench points lie in the grid, they are considered continental arc points, and their arc segment lengths are appended to the total continental arc length for the specifiedtime. The total length is scaled to kilometres using the geocentric Earth radius.Parameters
time:int- The geological time at which to calculate total continental arc lengths.
continental_grid:Raster, array_like,orstr- The continental grid used to identify continental arc points. Must
be convertible to
Raster. For an array, a global extent is assumed [-180,180,-90,90]. For a filename, the extent is obtained from the file. trench_arc_distance:float- The trench-to-arc distance (in kilometres) to project sampled trench points out by in the direction of their subduction polarities.
ignore_warnings:bool, default=True- Choose whether to ignore warning messages from PTT's subduction_convergence workflow that alerts the user of subduction sub-segments that are ignored due to unidentified polarities and/or subducting plates.
Returns
total_continental_arc_length_kms:float- The continental arc length (in km) at the specified time.
Expand source code
def total_continental_arc_length( self, time, continental_grid, trench_arc_distance, ignore_warnings=True, ): """Calculates the total length of all global continental arcs (km) at the specified geological time (Ma). Uses Plate Tectonic Tools' `subduction_convergence` workflow to sample a given plate model's trench features into point features and obtain their subduction polarities. The resolved points are projected out by the `trench_arc_distance` and their new locations are linearly interpolated onto the supplied `continental_grid`. If the projected trench points lie in the grid, they are considered continental arc points, and their arc segment lengths are appended to the total continental arc length for the specified `time`. The total length is scaled to kilometres using the geocentric Earth radius. Parameters ---------- time : int The geological time at which to calculate total continental arc lengths. continental_grid: Raster, array_like, or str The continental grid used to identify continental arc points. Must be convertible to `Raster`. For an array, a global extent is assumed [-180,180,-90,90]. For a filename, the extent is obtained from the file. trench_arc_distance : float The trench-to-arc distance (in kilometres) to project sampled trench points out by in the direction of their subduction polarities. ignore_warnings : bool, default=True Choose whether to ignore warning messages from PTT's subduction_convergence workflow that alerts the user of subduction sub-segments that are ignored due to unidentified polarities and/or subducting plates. Returns ------- total_continental_arc_length_kms : float The continental arc length (in km) at the specified time. """ from . import grids as _grids if isinstance(continental_grid, _grids.Raster): graster = continental_grid elif isinstance(continental_grid, str): # Process the continental grid directory graster = _grids.Raster( data=continental_grid, realign=True, time=float(time), ) else: # Process the masked continental grid try: continental_grid = np.array(continental_grid) graster = _grids.Raster( data=continental_grid, extent=[-180, 180, -90, 90], time=float(time), ) except Exception as e: raise TypeError( "Invalid type for `continental_grid` (must be Raster," + " str, or array_like)" ) from e if (time != graster.time) and (not ignore_warnings): raise RuntimeWarning( "`continental_grid.time` ({}) ".format(graster.time) + "does not match `time` ({})".format(time) ) # Obtain trench data with Plate Tectonic Tools trench_data = self.tessellate_subduction_zones( time, ignore_warnings=ignore_warnings ) # Extract trench data trench_normal_azimuthal_angle = trench_data[:, 7] trench_arcseg = trench_data[:, 6] trench_pt_lon = trench_data[:, 0] trench_pt_lat = trench_data[:, 1] # Modify the trench-arc distance using the geocentric radius arc_distance = trench_arc_distance / ( _tools.geocentric_radius(trench_pt_lat) / 1000 ) # Project trench points out along trench-arc distance, and obtain their new lat-lon coordinates dlon = arc_distance * np.sin(np.radians(trench_normal_azimuthal_angle)) dlat = arc_distance * np.cos(np.radians(trench_normal_azimuthal_angle)) ilon = trench_pt_lon + np.degrees(dlon) ilat = trench_pt_lat + np.degrees(dlat) # Linearly interpolate projected points onto continental grids, and collect the indices of points that lie # within the grids. sampled_points = graster.interpolate( ilon, ilat, method="linear", return_indices=False, ) continental_indices = np.where(sampled_points > 0) point_lats = ilat[continental_indices] point_radii = _tools.geocentric_radius(point_lats) * 1.0e-3 # km segment_arclens = np.deg2rad(trench_arcseg[continental_indices]) segment_lengths = point_radii * segment_arclens return np.sum(segment_lengths) def total_ridge_length(self, time, use_ptt=False, ignore_warnings=False)-
Calculates the total length of all mid-ocean ridges (km) at the specified geological time (Ma).
if
use_pttis TrueUses Plate Tectonic Tools'
ridge_spreading_rateworkflow to calculate ridge segment lengths. Scales lengths to kilometres using the geocentric radius.Otherwise
Resolves topology features of the PlateReconstruction model and extracts their shared boundary sections. The lengths of each GPML mid-ocean ridge shared boundary section are appended to the total ridge length. Scales lengths to kilometres using the geocentric radius.
Parameters
time:int- The geological time at which to calculate total mid-ocean ridge lengths.
use_ptt:bool, default=False- If set to
True, the PTT method is used. ignore_warnings:bool, default=False- Choose whether to ignore warning messages from PTT's
ridge_spreading_rateworkflow.
Raises
ValueError- If neither
use_pygplatesoruse_ptthave been set toTrue.
Returns
total_ridge_length_kms:float- The total length of global mid-ocean ridges (in kilometres) at the specified time.
Expand source code
def total_ridge_length(self, time, use_ptt=False, ignore_warnings=False): """Calculates the total length of all mid-ocean ridges (km) at the specified geological time (Ma). if `use_ptt` is True Uses Plate Tectonic Tools' `ridge_spreading_rate` workflow to calculate ridge segment lengths. Scales lengths to kilometres using the geocentric radius. Otherwise Resolves topology features of the PlateReconstruction model and extracts their shared boundary sections. The lengths of each GPML mid-ocean ridge shared boundary section are appended to the total ridge length. Scales lengths to kilometres using the geocentric radius. Parameters ---------- time : int The geological time at which to calculate total mid-ocean ridge lengths. use_ptt : bool, default=False If set to `True`, the PTT method is used. ignore_warnings : bool, default=False Choose whether to ignore warning messages from PTT's `ridge_spreading_rate` workflow. Raises ------ ValueError If neither `use_pygplates` or `use_ptt` have been set to `True`. Returns ------- total_ridge_length_kms : float The total length of global mid-ocean ridges (in kilometres) at the specified time. """ from . import ptt as _ptt if use_ptt is True: with warnings.catch_warnings(): warnings.simplefilter("ignore") ridge_data = self.tessellate_mid_ocean_ridges(time) ridge_arcseg = ridge_data[:, 3] ridge_pt_lat = ridge_data[:, 1] total_ridge_length_kms = 0 for i, segment in enumerate(ridge_arcseg): earth_radius = _tools.geocentric_radius(ridge_pt_lat[i]) / 1e3 total_ridge_length_kms += np.deg2rad(segment) * earth_radius return total_ridge_length_kms else: resolved_topologies = [] shared_boundary_sections = [] pygplates.resolve_topologies( self.topology_features, self.rotation_model, resolved_topologies, time, shared_boundary_sections, ) total_ridge_length_kms = 0.0 for shared_boundary_section in shared_boundary_sections: if ( shared_boundary_section.get_feature().get_feature_type() != pygplates.FeatureType.gpml_mid_ocean_ridge ): continue for ( shared_sub_segment ) in shared_boundary_section.get_shared_sub_segments(): clat, clon = ( shared_sub_segment.get_resolved_geometry() .get_centroid() .to_lat_lon() ) earth_radius = _tools.geocentric_radius(clat) / 1e3 total_ridge_length_kms += ( shared_sub_segment.get_resolved_geometry().get_arc_length() * earth_radius ) return total_ridge_length_kms def total_subduction_zone_length(self, time, use_ptt=False, ignore_warnings=False)-
Calculates the total length of all mid-ocean ridges (km) at the specified geological time (Ma).
if
use_pttis TrueUses Plate Tectonic Tools'
subduction_convergencemodule to calculate trench segment lengths on a unit sphere. The aggregated total subduction zone length is scaled to kilometres using the geocentric radius.Otherwise
Resolves topology features ascribed to the
PlateReconstructionmodel and extracts their shared boundary sections. The lengths of each trench boundary section are appended to the total subduction zone length. The total length is scaled to kilometres using a latitude-dependent (geocentric) Earth radius.Parameters
time:int- The geological time at which to calculate total mid-ocean ridge lengths.
use_ptt:bool, default=False- If set to
True, the PTT method is used. ignore_warnings:bool, default=False- Choose whether to ignore warning messages from PTT's
subduction_convergenceworkflow. These warnings alert the user when certain subduction sub-segments are ignored - this happens when the trench segments have unidentifiable subduction polarities and/or subducting plates.
Raises
ValueError- If neither
use_pygplatesoruse_ptthave been set toTrue.
Returns
total_subduction_zone_length_kms:float- The total subduction zone length (in km) at the specified
time.
Expand source code
def total_subduction_zone_length(self, time, use_ptt=False, ignore_warnings=False): """Calculates the total length of all mid-ocean ridges (km) at the specified geological time (Ma). if `use_ptt` is True Uses Plate Tectonic Tools' `subduction_convergence` module to calculate trench segment lengths on a unit sphere. The aggregated total subduction zone length is scaled to kilometres using the geocentric radius. Otherwise Resolves topology features ascribed to the `PlateReconstruction` model and extracts their shared boundary sections. The lengths of each trench boundary section are appended to the total subduction zone length. The total length is scaled to kilometres using a latitude-dependent (geocentric) Earth radius. Parameters ---------- time : int The geological time at which to calculate total mid-ocean ridge lengths. use_ptt : bool, default=False If set to `True`, the PTT method is used. ignore_warnings : bool, default=False Choose whether to ignore warning messages from PTT's `subduction_convergence` workflow. These warnings alert the user when certain subduction sub-segments are ignored - this happens when the trench segments have unidentifiable subduction polarities and/or subducting plates. Raises ------ ValueError If neither `use_pygplates` or `use_ptt` have been set to `True`. Returns ------- total_subduction_zone_length_kms : float The total subduction zone length (in km) at the specified `time`. """ from . import ptt as _ptt if use_ptt: with warnings.catch_warnings(): warnings.simplefilter("ignore") subduction_data = self.tessellate_subduction_zones( time, ignore_warnings=ignore_warnings ) trench_arcseg = subduction_data[:, 6] trench_pt_lat = subduction_data[:, 1] total_subduction_zone_length_kms = 0 for i, segment in enumerate(trench_arcseg): earth_radius = _tools.geocentric_radius(trench_pt_lat[i]) / 1e3 total_subduction_zone_length_kms += np.deg2rad(segment) * earth_radius return total_subduction_zone_length_kms else: resolved_topologies = [] shared_boundary_sections = [] pygplates.resolve_topologies( self.topology_features, self.rotation_model, resolved_topologies, time, shared_boundary_sections, ) total_subduction_zone_length_kms = 0.0 for shared_boundary_section in shared_boundary_sections: if ( shared_boundary_section.get_feature().get_feature_type() != pygplates.FeatureType.gpml_subduction_zone ): continue for ( shared_sub_segment ) in shared_boundary_section.get_shared_sub_segments(): clat, clon = ( shared_sub_segment.get_resolved_geometry() .get_centroid() .to_lat_lon() ) earth_radius = _tools.geocentric_radius(clat) / 1e3 total_subduction_zone_length_kms += ( shared_sub_segment.get_resolved_geometry().get_arc_length() * earth_radius ) return total_subduction_zone_length_kms
class PlotTopologies (plate_reconstruction, coastlines=None, continents=None, COBs=None, time=None, anchor_plate_id=0)-
A class with tools to read, reconstruct and plot topology features at specific reconstruction times.
PlotTopologiesis a shorthand for PyGPlates and Shapely functionalities that:- Read features held in GPlates GPML (GPlates Markup Language) files and ESRI shapefiles;
- Reconstruct the locations of these features as they migrate through geological time;
- Turn these reconstructed features into Shapely geometries for plotting
on
cartopy.mpl.geoaxes.GeoAxesorcartopy.mpl.geoaxes.GeoAxesSubplotmap Projections.
To call the
PlotTopologiesobject, supply:- an instance of the GPlately
plate_reconstructionobject
and optionally,
- a
coastline_filename - a
continent_filename - a
COB_filename - a reconstruction
time - an
anchor_plate_id
For example:
# Calling the PlotTopologies object gplot = gplately.plot.PlotTopologies(plate_reconstruction, coastline_filename=None, continent_filename=None, COB_filename=None, time=None, anchor_plate_id=0, ) # Setting a new reconstruction time gplot.time = 20 # MaThe
coastline_filename,continent_filenameandCOB_filenamecan be single strings to GPML and/or shapefiles, as well as instances ofFeatureCollection. If using GPlately'sDataServerobject to source these files, they will be passed asFeatureCollectionitems.Some features for plotting (like plate boundaries) are taken from the
PlateReconstructionobject'stopology_featuresattribute. They have already been reconstructed to the giventimeusing Plate Tectonic Tools. Simply provide a new reconstruction time by changing thetimeattribute, e.g.gplot.time = 20 # Mawhich will automatically reconstruct all topologies to the specified time. You MUST set
gplot.timebefore plotting anything.A variety of geological features can be plotted on GeoAxes/GeoAxesSubplot maps as Shapely
MultiLineStringorMultiPolygongeometries, including:- subduction boundaries & subduction polarity teeth
- mid-ocean ridge boundaries
- transform boundaries
- miscellaneous boundaries
- coastline polylines
- continental polygons and
- continent-ocean boundary polylines
- topological plate velocity vector fields
- netCDF4 MaskedArray or ndarray raster data:
- seafloor age grids
- paleo-age grids
- global relief (topography and bathymetry)
- assorted reconstructable feature data, for example:
- seafloor fabric
- large igneous provinces
- volcanic provinces
Attributes
plate_reconstruction:instanceof<gplately.reconstruction.PlateReconstruction>- The GPlately
PlateReconstructionobject will be used to access a platerotation_modeland a set oftopology_featureswhich contains plate boundary features like trenches, ridges and transforms. anchor_plate_id:int, default0- The anchor plate ID used for reconstruction.
base_projection:instanceof<cartopy.crs.{transform}>, default<cartopy.crs.PlateCarree> object- where {transform} is the map Projection to use on the Cartopy GeoAxes. By default, the base projection is set to cartopy.crs.PlateCarree. See the Cartopy projection list for all supported Projection types.
coastlines:str,orinstanceof<pygplates.FeatureCollection>- The full string path to a coastline feature file. Coastline features can also
be passed as instances of the
FeatureCollectionobject (this is the case if these features are sourced from theDataServerobject). continents:str,orinstanceof<pygplates.FeatureCollection>- The full string path to a continent feature file. Continent features can also
be passed as instances of the
FeatureCollectionobject (this is the case if these features are sourced from theDataServerobject). COBs:str,orinstanceof<pygplates.FeatureCollection>- The full string path to a COB feature file. COB features can also be passed
as instances of the
FeatureCollectionobject (this is the case if these features are sourced from theDataServerobject). coastlines:iterable/listof<pygplates.ReconstructedFeatureGeometry>- A list containing coastline features reconstructed to the specified
timeattribute. continents:iterable/listof<pygplates.ReconstructedFeatureGeometry>- A list containing continent features reconstructed to the specified
timeattribute. COBs:iterable/listof<pygplates.ReconstructedFeatureGeometry>- A list containing COB features reconstructed to the specified
timeattribute. time:float- The time (Ma) to reconstruct and plot geological features to.
topologies:iterable/listof<pygplates.Feature>-
A list containing assorted topologies like:
- pygplates.FeatureType.gpml_topological_network
- pygplates.FeatureType.gpml_oceanic_crust
- pygplates.FeatureType.gpml_topological_slab_boundary
- pygplates.FeatureType.gpml_topological_closed_plate_boundary
ridge_transforms:iterable/listof<pygplates.Feature>- A list containing ridge and transform boundary sections of type pygplates.FeatureType.gpml_mid_ocean_ridge
ridges:iterable/listof<pygplates.Feature>- A list containing ridge boundary sections of type pygplates.FeatureType.gpml_mid_ocean_ridge
transforms:iterable/listof<pygplates.Feature>- A list containing transform boundary sections of type pygplates.FeatureType.gpml_mid_ocean_ridge
trenches:iterable/listof<pygplates.Feature>- A list containing trench boundary sections of type pygplates.FeatureType.gpml_subduction_zone
trench_left:iterable/listof<pygplates.Feature>- A list containing left subduction boundary sections of type pygplates.FeatureType.gpml_subduction_zone
trench_right:iterable/listof<pygplates.Feature>- A list containing right subduction boundary sections of type pygplates.FeatureType.gpml_subduction_zone
other:iterable/listof<pygplates.Feature>- A list containing other geological features like unclassified features, extended continental crusts, continental rifts, faults, orogenic belts, fracture zones, inferred paleo boundaries, terrane boundaries and passive continental boundaries.
Expand source code
class PlotTopologies(object): """A class with tools to read, reconstruct and plot topology features at specific reconstruction times. `PlotTopologies` is a shorthand for PyGPlates and Shapely functionalities that: * Read features held in GPlates GPML (GPlates Markup Language) files and ESRI shapefiles; * Reconstruct the locations of these features as they migrate through geological time; * Turn these reconstructed features into Shapely geometries for plotting on `cartopy.mpl.geoaxes.GeoAxes` or `cartopy.mpl.geoaxes.GeoAxesSubplot` map Projections. To call the `PlotTopologies` object, supply: * an instance of the GPlately `plate_reconstruction` object and optionally, * a `coastline_filename` * a `continent_filename` * a `COB_filename` * a reconstruction `time` * an `anchor_plate_id` For example: # Calling the PlotTopologies object gplot = gplately.plot.PlotTopologies(plate_reconstruction, coastline_filename=None, continent_filename=None, COB_filename=None, time=None, anchor_plate_id=0, ) # Setting a new reconstruction time gplot.time = 20 # Ma The `coastline_filename`, `continent_filename` and `COB_filename` can be single strings to GPML and/or shapefiles, as well as instances of `pygplates.FeatureCollection`. If using GPlately's `DataServer` object to source these files, they will be passed as `pygplates.FeatureCollection` items. Some features for plotting (like plate boundaries) are taken from the `PlateReconstruction` object's`topology_features` attribute. They have already been reconstructed to the given `time` using [Plate Tectonic Tools](https://github.com/EarthByte/PlateTectonicTools). Simply provide a new reconstruction time by changing the `time` attribute, e.g. gplot.time = 20 # Ma which will automatically reconstruct all topologies to the specified time. You __MUST__ set `gplot.time` before plotting anything. A variety of geological features can be plotted on GeoAxes/GeoAxesSubplot maps as Shapely `MultiLineString` or `MultiPolygon` geometries, including: * subduction boundaries & subduction polarity teeth * mid-ocean ridge boundaries * transform boundaries * miscellaneous boundaries * coastline polylines * continental polygons and * continent-ocean boundary polylines * topological plate velocity vector fields * netCDF4 MaskedArray or ndarray raster data: - seafloor age grids - paleo-age grids - global relief (topography and bathymetry) * assorted reconstructable feature data, for example: - seafloor fabric - large igneous provinces - volcanic provinces Attributes ---------- plate_reconstruction : instance of <gplately.reconstruction.PlateReconstruction> The GPlately `PlateReconstruction` object will be used to access a plate `rotation_model` and a set of `topology_features` which contains plate boundary features like trenches, ridges and transforms. anchor_plate_id : int, default 0 The anchor plate ID used for reconstruction. base_projection : instance of <cartopy.crs.{transform}>, default <cartopy.crs.PlateCarree> object where {transform} is the map Projection to use on the Cartopy GeoAxes. By default, the base projection is set to cartopy.crs.PlateCarree. See the [Cartopy projection list](https://scitools.org.uk/cartopy/docs/v0.15/crs/projections.html) for all supported Projection types. coastlines : str, or instance of <pygplates.FeatureCollection> The full string path to a coastline feature file. Coastline features can also be passed as instances of the `pygplates.FeatureCollection` object (this is the case if these features are sourced from the `DataServer` object). continents : str, or instance of <pygplates.FeatureCollection> The full string path to a continent feature file. Continent features can also be passed as instances of the `pygplates.FeatureCollection` object (this is the case if these features are sourced from the `DataServer` object). COBs : str, or instance of <pygplates.FeatureCollection> The full string path to a COB feature file. COB features can also be passed as instances of the `pygplates.FeatureCollection` object (this is the case if these features are sourced from the `DataServer` object). coastlines : iterable/list of <pygplates.ReconstructedFeatureGeometry> A list containing coastline features reconstructed to the specified `time` attribute. continents : iterable/list of <pygplates.ReconstructedFeatureGeometry> A list containing continent features reconstructed to the specified `time` attribute. COBs : iterable/list of <pygplates.ReconstructedFeatureGeometry> A list containing COB features reconstructed to the specified `time` attribute. time : float The time (Ma) to reconstruct and plot geological features to. topologies : iterable/list of <pygplates.Feature> A list containing assorted topologies like: - pygplates.FeatureType.gpml_topological_network - pygplates.FeatureType.gpml_oceanic_crust - pygplates.FeatureType.gpml_topological_slab_boundary - pygplates.FeatureType.gpml_topological_closed_plate_boundary ridge_transforms : iterable/list of <pygplates.Feature> A list containing ridge and transform boundary sections of type pygplates.FeatureType.gpml_mid_ocean_ridge ridges : iterable/list of <pygplates.Feature> A list containing ridge boundary sections of type pygplates.FeatureType.gpml_mid_ocean_ridge transforms : iterable/list of <pygplates.Feature> A list containing transform boundary sections of type pygplates.FeatureType.gpml_mid_ocean_ridge trenches : iterable/list of <pygplates.Feature> A list containing trench boundary sections of type pygplates.FeatureType.gpml_subduction_zone trench_left : iterable/list of <pygplates.Feature> A list containing left subduction boundary sections of type pygplates.FeatureType.gpml_subduction_zone trench_right : iterable/list of <pygplates.Feature> A list containing right subduction boundary sections of type pygplates.FeatureType.gpml_subduction_zone other : iterable/list of <pygplates.Feature> A list containing other geological features like unclassified features, extended continental crusts, continental rifts, faults, orogenic belts, fracture zones, inferred paleo boundaries, terrane boundaries and passive continental boundaries. """ def __init__( self, plate_reconstruction, coastlines=None, continents=None, COBs=None, time=None, anchor_plate_id=0, ): self.plate_reconstruction = plate_reconstruction if self.plate_reconstruction.topology_features is None: raise ValueError("Plate model must have topology features.") self.base_projection = ccrs.PlateCarree() # store these for when time is updated # make sure these are initialised as FeatureCollection objects self._coastlines = _load_FeatureCollection(coastlines) self._continents = _load_FeatureCollection(continents) self._COBs = _load_FeatureCollection(COBs) self.coastlines = None self.continents = None self.COBs = None self._anchor_plate_id = self._check_anchor_plate_id(anchor_plate_id) # store topologies for easy access # setting time runs the update_time routine self._time = None if time is not None: self.time = time def __getstate__(self): filenames = self.plate_reconstruction.__getstate__() # add important variables from Points object if self._coastlines: filenames["coastlines"] = self._coastlines.filenames if self._continents: filenames["continents"] = self._continents.filenames if self._COBs: filenames["COBs"] = self._COBs.filenames filenames["time"] = self.time filenames["plate_id"] = self._anchor_plate_id return filenames def __setstate__(self, state): plate_reconstruction_args = [state["rotation_model"], None, None] if "topology_features" in state: plate_reconstruction_args[1] = state["topology_features"] if "static_polygons" in state: plate_reconstruction_args[2] = state["static_polygons"] self.plate_reconstruction = _PlateReconstruction(*plate_reconstruction_args) self._coastlines = None self._continents = None self._COBs = None self.coastlines = None self.continents = None self.COBs = None # reinstate unpicklable items if "coastlines" in state: self._coastlines = _FeatureCollection() for feature in state["coastlines"]: self._coastlines.add(_FeatureCollection(feature)) if "continents" in state: self._continents = _FeatureCollection() for feature in state["continents"]: self._continents.add(_FeatureCollection(feature)) if "COBs" in state: self._COBs = _FeatureCollection() for feature in state["COBs"]: self._COBs.add(_FeatureCollection(feature)) self._anchor_plate_id = state["plate_id"] self.base_projection = ccrs.PlateCarree() self._time = None @property def time(self): """The reconstruction time.""" return self._time @time.setter def time(self, var): """Allows the time attribute to be changed. Updates all instances of the time attribute in the object (e.g. reconstructions and resolving topologies will use this new time). Raises ------ ValueError If the chosen reconstruction time is <0 Ma. """ if var == self.time: pass elif var >= 0: self.update_time(var) else: raise ValueError("Enter a valid time >= 0") @property def anchor_plate_id(self): """Anchor plate ID for reconstruction. Must be an integer >= 0.""" return self._anchor_plate_id @anchor_plate_id.setter def anchor_plate_id(self, anchor_plate): self._anchor_plate_id = self._check_anchor_plate_id(anchor_plate) self.update_time(self.time) @staticmethod def _check_anchor_plate_id(id): id = int(id) if id < 0: raise ValueError("Invalid anchor plate ID: {}".format(id)) return id def update_time(self, time): """Re-reconstruct features and topologies to the time specified by the `PlotTopologies` `time` attribute whenever it or the anchor plate is updated. Notes ----- The following `PlotTopologies` attributes are updated whenever a reconstruction `time` attribute is set: - resolved topology features (topological plates and networks) - ridge and transform boundary sections (resolved features) - ridge boundary sections (resolved features) - transform boundary sections (resolved features) - subduction boundary sections (resolved features) - left subduction boundary sections (resolved features) - right subduction boundary sections (resolved features) - other boundary sections (resolved features) that are not subduction zones or mid-ocean ridges (ridge/transform) Moreover, coastlines, continents and COBs are reconstructed to the new specified `time`. """ self._time = float(time) resolved_topologies = ptt.resolve_topologies.resolve_topologies_into_features( self.plate_reconstruction.rotation_model, self.plate_reconstruction.topology_features, self.time, ) ( self.topologies, self.ridge_transforms, self.ridges, self.transforms, self.trenches, self.trench_left, self.trench_right, self.other, ) = resolved_topologies # miscellaneous boundaries self.continental_rifts = [] self.faults = [] self.fracture_zones = [] self.inferred_paleo_boundaries = [] self.terrane_boundaries = [] self.transitional_crusts = [] self.orogenic_belts = [] self.sutures = [] self.continental_crusts = [] self.extended_continental_crusts = [] self.passive_continental_boundaries = [] self.slab_edges = [] self.misc_transforms = [] self.unclassified_features = [] for topol in self.other: if topol.get_feature_type() == pygplates.FeatureType.gpml_continental_rift: self.continental_rifts.append(topol) elif topol.get_feature_type() == pygplates.FeatureType.gpml_fault: self.faults.append(topol) elif topol.get_feature_type() == pygplates.FeatureType.gpml_fracture_zone: self.fracture_zones.append(topol) elif ( topol.get_feature_type() == pygplates.FeatureType.gpml_inferred_paleo_boundary ): self.inferred_paleo_boundaries.append(topol) elif ( topol.get_feature_type() == pygplates.FeatureType.gpml_terrane_boundary ): self.terrane_boundaries.append(topol) elif ( topol.get_feature_type() == pygplates.FeatureType.gpml_transitional_crust ): self.transitional_crusts.append(topol) elif topol.get_feature_type() == pygplates.FeatureType.gpml_orogenic_belt: self.orogenic_belts.append(topol) elif topol.get_feature_type() == pygplates.FeatureType.gpml_suture: self.sutures.append(topol) elif ( topol.get_feature_type() == pygplates.FeatureType.gpml_continental_crust ): self.continental_crusts.append(topol) elif ( topol.get_feature_type() == pygplates.FeatureType.gpml_extended_continental_crust ): self.extended_continental_crusts.append(topol) elif ( topol.get_feature_type() == pygplates.FeatureType.gpml_passive_continental_boundary ): self.passive_continental_boundaries.append(topol) elif topol.get_feature_type() == pygplates.FeatureType.gpml_slab_edge: self.slab_edges.append(topol) elif topol.get_feature_type() == pygplates.FeatureType.gpml_transform: self.misc_transforms.append(topol) elif ( topol.get_feature_type() == pygplates.FeatureType.gpml_unclassified_feature ): self.unclassified_features.append(topol) # reconstruct other important polygons and lines if self._coastlines: self.coastlines = self.plate_reconstruction.reconstruct( self._coastlines, self.time, from_time=0, anchor_plate_id=self.anchor_plate_id, ) if self._continents: self.continents = self.plate_reconstruction.reconstruct( self._continents, self.time, from_time=0, anchor_plate_id=self.anchor_plate_id, ) if self._COBs: self.COBs = self.plate_reconstruction.reconstruct( self._COBs, self.time, from_time=0, anchor_plate_id=self.anchor_plate_id ) # subduction teeth def _tessellate_triangles( self, features, tesselation_radians, triangle_base_length, triangle_aspect=1.0 ): """Places subduction teeth along subduction boundary line segments within a MultiLineString shapefile. Parameters ---------- shapefilename : str Path to shapefile containing the subduction boundary features tesselation_radians : float Parametrises subduction teeth density. Triangles are generated only along line segments with distances that exceed the given threshold tessellation_radians. triangle_base_length : float Length of teeth triangle base triangle_aspect : float, default=1.0 Aspect ratio of teeth triangles. Ratio is 1.0 by default. Returns ------- X_points : (n,3) array X points that define the teeth triangles Y_points : (n,3) array Y points that define the teeth triangles """ tesselation_degrees = np.degrees(tesselation_radians) triangle_pointsX = [] triangle_pointsY = [] date_line_wrapper = pygplates.DateLineWrapper() for feature in features: cum_distance = 0.0 for geometry in feature.get_geometries(): wrapped_lines = date_line_wrapper.wrap(geometry) for line in wrapped_lines: pts = np.array( [ (p.get_longitude(), p.get_latitude()) for p in line.get_points() ] ) for p in range(0, len(pts) - 1): A = pts[p] B = pts[p + 1] AB_dist = B - A AB_norm = AB_dist / np.hypot(*AB_dist) cum_distance += np.hypot(*AB_dist) # create a new triangle if cumulative distance is exceeded. if cum_distance >= tesselation_degrees: C = A + triangle_base_length * AB_norm # find normal vector AD_dist = np.array([AB_norm[1], -AB_norm[0]]) AD_norm = AD_dist / np.linalg.norm(AD_dist) C0 = A + 0.5 * triangle_base_length * AB_norm # project point along normal vector D = C0 + triangle_base_length * triangle_aspect * AD_norm triangle_pointsX.append([A[0], C[0], D[0]]) triangle_pointsY.append([A[1], C[1], D[1]]) cum_distance = 0.0 return np.array(triangle_pointsX), np.array(triangle_pointsY) def get_feature( self, feature, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed features. Notes ----- The feature needed to produce the GeoDataFrame should already be constructed to a `time`. This function converts the feature into a set of Shapely geometries whose coordinates are passed to a geopandas GeoDataFrame. Parameters ---------- feature : instance of <pygplates.Feature> A feature reconstructed to `time`. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `feature` geometries. """ shp = shapelify_features( feature, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": shp}, geometry="geometry") return gdf def plot_feature(self, ax, feature, **kwargs): """Plot pygplates.FeatureCollection or pygplates.Feature onto a map. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. **kwargs : Keyword arguments for parameters such as `facecolor`, `alpha`, etc. for plotting coastline geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with coastline features plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_feature( feature, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, **kwargs) def get_coastlines(self, central_meridian=0.0, tessellate_degrees=None): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed coastline polygons. Notes ----- The `coastlines` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `coastlines` are reconstructed, they are converted into Shapely polygons whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `coastlines` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `coastlines` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No coastlines have been resolved. Set `PlotTopologies.time` to construct coastlines." ) if self.coastlines is None: raise ValueError("Supply coastlines to PlotTopologies object") coastline_polygons = shapelify_feature_polygons( self.coastlines, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": coastline_polygons}, geometry="geometry") return gdf def plot_coastlines(self, ax, **kwargs): """Plot reconstructed coastline polygons onto a standard map Projection. Notes ----- The `coastlines` for plotting are accessed from the `PlotTopologies` object's `coastlines` attribute. These `coastlines` are reconstructed to the `time` passed to the `PlotTopologies` object and converted into Shapely polylines. The reconstructed `coastlines` are added onto the GeoAxes or GeoAxesSubplot map `ax` using GeoPandas. Map resentation details (e.g. facecolor, edgecolor, alpha…) are permitted as keyword arguments. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. **kwargs : Keyword arguments for parameters such as `facecolor`, `alpha`, etc. for plotting coastline geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with coastline features plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_coastlines( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, **kwargs) def get_continents(self, central_meridian=0.0, tessellate_degrees=None): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed continental polygons. Notes ----- The `continents` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `continents` are reconstructed, they are converted into Shapely polygons whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `continents` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `continents` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No continents have been resolved. Set `PlotTopologies.time` to construct continents." ) if self.continents is None: raise ValueError("Supply continents to PlotTopologies object") continent_polygons = shapelify_feature_polygons( self.continents, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": continent_polygons}, geometry="geometry") return gdf def plot_continents(self, ax, **kwargs): """Plot reconstructed continental polygons onto a standard map Projection. Notes ----- The `continents` for plotting are accessed from the `PlotTopologies` object's `continents` attribute. These `continents` are reconstructed to the `time` passed to the `PlotTopologies` object and converted into Shapely polygons. The reconstructed `coastlines` are plotted onto the GeoAxes or GeoAxesSubplot map `ax` using GeoPandas. Map presentation details (e.g. facecolor, edgecolor, alpha…) are permitted as keyword arguments. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. **kwargs : Keyword arguments for parameters such as `facecolor`, `alpha`, etc. for plotting continental geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with continent features plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_continents( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, **kwargs) def get_continent_ocean_boundaries( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed continent-ocean boundary lines. Notes ----- The `COBs` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `COBs` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `COBs` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `COBs` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No geometries have been resolved. Set `PlotTopologies.time` to construct topologies." ) if self.COBs is None: raise ValueError("Supply COBs to PlotTopologies object") COB_lines = shapelify_feature_lines( self.COBs, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": COB_lines}, geometry="geometry") return gdf def plot_continent_ocean_boundaries(self, ax, **kwargs): """Plot reconstructed continent-ocean boundary (COB) polygons onto a standard map Projection. Notes ----- The `COBs` for plotting are accessed from the `PlotTopologies` object's `COBs` attribute. These `COBs` are reconstructed to the `time` passed to the `PlotTopologies` object and converted into Shapely polylines. The reconstructed `COBs` are plotted onto the GeoAxes or GeoAxesSubplot map `ax` using GeoPandas. Map presentation details (e.g. `facecolor`, `edgecolor`, `alpha`…) are permitted as keyword arguments. These COBs are transformed into shapely geometries and added onto the chosen map for a specific geological time (supplied to the PlotTopologies object). Map presentation details (e.g. facecolor, edgecolor, alpha…) are permitted. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. **kwargs : Keyword arguments for parameters such as `facecolor`, `alpha`, etc. for plotting COB geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with COB features plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_continent_ocean_boundaries( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, **kwargs) def get_ridges(self, central_meridian=0.0, tessellate_degrees=None): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed ridge lines. Notes ----- The `ridges` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `ridges` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `ridges` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `ridges` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No ridges have been resolved. Set `PlotTopologies.time` to construct ridges." ) if self.ridges is None: raise ValueError("No ridge topologies passed to PlotTopologies.") ridge_lines = shapelify_feature_lines( self.ridges, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": ridge_lines}, geometry="geometry") return gdf def plot_ridges(self, ax, color="black", **kwargs): """Plot reconstructed ridge polylines onto a standard map Projection. Notes ----- The `ridges` for plotting are accessed from the `PlotTopologies` object's `ridges` attribute. These `ridges` are reconstructed to the `time` passed to the `PlotTopologies` object and converted into Shapely polylines. The reconstructed `ridges` are plotted onto the GeoAxes or GeoAxesSubplot map `ax` using GeoPandas. Map presentation details (e.g. `facecolor`, `edgecolor`, `alpha`…) are permitted as keyword arguments. Ridge geometries are wrapped to the dateline using pyGPlates' [DateLineWrapper](https://www.gplates.org/docs/pygplates/generated/pygplates.datelinewrapper) by splitting a polyline into multiple polylines at the dateline. This is to avoid horizontal lines being formed between polylines at longitudes of -180 and 180 degrees. Point features near the poles (-89 & 89 degree latitude) are also clipped to ensure compatibility with Cartopy. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the ridge lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting ridge geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with ridge features plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_ridges( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def get_ridges_and_transforms( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed ridge and transform lines. Notes ----- The `ridge_transforms` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `ridge_transforms` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `ridges` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `ridge_transforms` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No ridges and transforms have been resolved. Set `PlotTopologies.time` to construct ridges and transforms." ) if self.ridge_transforms is None: raise ValueError( "No ridge and transform topologies passed to PlotTopologies." ) ridge_transform_lines = shapelify_feature_lines( self.ridge_transforms, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": ridge_transform_lines}, geometry="geometry") return gdf def plot_ridges_and_transforms(self, ax, color="black", **kwargs): """Plot reconstructed ridge & transform boundary polylines onto a standard map Projection. Notes ----- The ridge & transform sections for plotting are accessed from the `PlotTopologies` object's `ridge_transforms` attribute. These `ridge_transforms` are reconstructed to the `time` passed to the `PlotTopologies` object and converted into Shapely polylines. The reconstructed `ridge_transforms` are plotted onto the GeoAxes or GeoAxesSubplot map `ax` using GeoPandas. Map presentation details (e.g. `facecolor`, `edgecolor`, `alpha`…) are permitted as keyword arguments. Note: Ridge & transform geometries are wrapped to the dateline using pyGPlates' [DateLineWrapper](https://www.gplates.org/docs/pygplates/generated/pygplates.datelinewrapper) by splitting a polyline into multiple polylines at the dateline. This is to avoid horizontal lines being formed between polylines at longitudes of -180 and 180 degrees. Point features near the poles (-89 & 89 degree latitude) are also clipped to ensure compatibility with Cartopy. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the ridge & transform lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as ‘alpha’, etc. for plotting ridge & transform geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with ridge & transform features plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_ridges_and_transforms( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def get_transforms(self, central_meridian=0.0, tessellate_degrees=None): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed transform lines. Notes ----- The `transforms` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `transforms` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `transforms` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `transforms` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No transforms have been resolved. Set `PlotTopologies.time` to construct transforms." ) if self.transforms is None: raise ValueError("No transform topologies passed to PlotTopologies.") transform_lines = shapelify_feature_lines( self.transforms, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": transform_lines}, geometry="geometry") return gdf def plot_transforms(self, ax, color="black", **kwargs): """Plot reconstructed transform boundary polylines onto a standard map. Notes ----- The transform sections for plotting are accessed from the `PlotTopologies` object's `transforms` attribute. These `transforms` are reconstructed to the `time` passed to the `PlotTopologies` object and converted into Shapely polylines. The reconstructed `transforms` are plotted onto the GeoAxes or GeoAxesSubplot map `ax` using GeoPandas. Map presentation details (e.g. `facecolor`, `edgecolor`, `alpha`…) are permitted as keyword arguments. Transform geometries are wrapped to the dateline using pyGPlates' [DateLineWrapper](https://www.gplates.org/docs/pygplates/generated/pygplates.datelinewrapper) by splitting a polyline into multiple polylines at the dateline. This is to avoid horizontal lines being formed between polylines at longitudes of -180 and 180 degrees. Point features near the poles (-89 & 89 degree latitude) are also clipped to ensure compatibility with Cartopy. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the transform lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting transform geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with transform features plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_transforms( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def get_trenches(self, central_meridian=0.0, tessellate_degrees=None): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed trench lines. Notes ----- The `trenches` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `trenches` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `trenches` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `trenches` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No trenches have been resolved. Set `PlotTopologies.time` to construct trenches." ) if self.trenches is None: raise ValueError("No trenches passed to PlotTopologies.") trench_lines = shapelify_feature_lines( self.trenches, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": trench_lines}, geometry="geometry") return gdf def plot_trenches(self, ax, color="black", **kwargs): """Plot reconstructed subduction trench polylines onto a standard map Projection. Notes ----- The trench sections for plotting are accessed from the `PlotTopologies` object's `trenches` attribute. These `trenches` are reconstructed to the `time` passed to the `PlotTopologies` object and converted into Shapely polylines. The reconstructed `trenches` are plotted onto the GeoAxes or GeoAxesSubplot map `ax` using GeoPandas. Map presentation details (e.g. `facecolor`, `edgecolor`, `alpha`…) are permitted as keyword arguments. Trench geometries are wrapped to the dateline using pyGPlates' [DateLineWrapper](https://www.gplates.org/docs/pygplates/generated/pygplates.datelinewrapper) by splitting a polyline into multiple polylines at the dateline. This is to avoid horizontal lines being formed between polylines at longitudes of -180 and 180 degrees. Point features near the poles (-89 & 89 degree latitude) are also clipped to ensure compatibility with Cartopy. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with transform features plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_trenches( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def get_misc_boundaries(self, central_meridian=0.0, tessellate_degrees=None): """Create a geopandas.GeoDataFrame object containing geometries of other reconstructed lines. Notes ----- The `other` geometries needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `other` geometries are reconstructed, they are converted into Shapely features whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `other` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `other` geometries to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No miscellaneous topologies have been resolved. Set `PlotTopologies.time` to construct them." ) if self.other is None: raise ValueError("No miscellaneous topologies passed to PlotTopologies.") lines = shapelify_features( self.other, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": lines}, geometry="geometry") return gdf def plot_misc_boundaries(self, ax, color="black", **kwargs): """Plot reconstructed miscellaneous plate boundary polylines onto a standard map Projection. Notes ----- The miscellaneous boundary sections for plotting are accessed from the `PlotTopologies` object's `other` attribute. These `other` boundaries are reconstructed to the `time` passed to the `PlotTopologies` object and converted into Shapely polylines. The reconstructed `other` boundaries are plotted onto the GeoAxes or GeoAxesSubplot map `ax` using GeoPandas. Map presentation details (e.g. `facecolor`, `edgecolor`, `alpha`…) are permitted as keyword arguments. Miscellaneous boundary geometries are wrapped to the dateline using pyGPlates' [DateLineWrapper](https://www.gplates.org/docs/pygplates/generated/pygplates.datelinewrapper) by splitting a polyline into multiple polylines at the dateline. This is to avoid horizontal lines being formed between polylines at longitudes of -180 and 180 degrees. Point features near the poles (-89 & 89 degree latitude) are also clipped to ensure compatibility with Cartopy. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the boundary lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as ‘alpha’, etc. for plotting miscellaneous boundary geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with miscellaneous boundary features plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_misc_boundaries( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def plot_subduction_teeth_deprecated( self, ax, spacing=0.1, size=2.0, aspect=1, color="black", **kwargs ): """Plot subduction teeth onto a standard map Projection. Notes ----- Subduction teeth are tessellated from `PlotTopologies` object attributes `trench_left` and `trench_right`, and transformed into Shapely polygons for plotting. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. spacing : float, default=0.1 The tessellation threshold (in radians). Parametrises subduction tooth density. Triangles are generated only along line segments with distances that exceed the given threshold ‘spacing’. size : float, default=2.0 Length of teeth triangle base. aspect : float, default=1 Aspect ratio of teeth triangles. Ratio is 1.0 by default. color : str, default=’black’ The colour of the teeth. By default, it is set to black. **kwargs : Keyword arguments for parameters such as ‘alpha’, etc. for plotting subduction tooth polygons. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with subduction teeth plotted onto the chosen map projection. """ import shapely # add Subduction Teeth subd_xL, subd_yL = self._tessellate_triangles( self.trench_left, tesselation_radians=spacing, triangle_base_length=size, triangle_aspect=-aspect, ) subd_xR, subd_yR = self._tessellate_triangles( self.trench_right, tesselation_radians=spacing, triangle_base_length=size, triangle_aspect=aspect, ) teeth = [] for tX, tY in zip(subd_xL, subd_yL): triangle_xy_points = np.c_[tX, tY] shp = shapely.geometry.Polygon(triangle_xy_points) teeth.append(shp) for tX, tY in zip(subd_xR, subd_yR): triangle_xy_points = np.c_[tX, tY] shp = shapely.geometry.Polygon(triangle_xy_points) teeth.append(shp) return ax.add_geometries(teeth, crs=self.base_projection, color=color, **kwargs) def get_subduction_direction(self): """Create a geopandas.GeoDataFrame object containing geometries of trench directions. Notes ----- The `trench_left` and `trench_right` geometries needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `other` geometries are reconstructed, they are converted into Shapely features whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf_left : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `trench_left` geometry. gdf_right : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `trench_right` geometry. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `trench_left` or `trench_right` geometries to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No miscellaneous topologies have been resolved. Set `PlotTopologies.time` to construct them." ) if self.trench_left is None or self.trench_right is None: raise ValueError( "No trench_left or trench_right topologies passed to PlotTopologies." ) trench_left_features = shapelify_feature_lines(self.trench_left) trench_right_features = shapelify_feature_lines(self.trench_right) gdf_left = gpd.GeoDataFrame( {"geometry": trench_left_features}, geometry="geometry" ) gdf_right = gpd.GeoDataFrame( {"geometry": trench_right_features}, geometry="geometry" ) return gdf_left, gdf_right def plot_subduction_teeth( self, ax, spacing=0.07, size=None, aspect=None, color="black", **kwargs ): """Plot subduction teeth onto a standard map Projection. Notes ----- Subduction teeth are tessellated from `PlotTopologies` object attributes `trench_left` and `trench_right`, and transformed into Shapely polygons for plotting. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. spacing : float, default=0.07 The tessellation threshold (in radians). Parametrises subduction tooth density. Triangles are generated only along line segments with distances that exceed the given threshold `spacing`. size : float, default=None Length of teeth triangle base (in radians). If kept at `None`, then `size = 0.5*spacing`. aspect : float, default=None Aspect ratio of teeth triangles. If kept at `None`, then `aspect = 2/3*size`. color : str, default='black' The colour of the teeth. By default, it is set to black. **kwargs : Keyword arguments parameters such as `alpha`, etc. for plotting subduction tooth polygons. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). """ if self._time is None: raise ValueError( "No topologies have been resolved. Set `PlotTopologies.time` to construct them." ) if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") central_meridian = _meridian_from_ax(ax) tessellate_degrees = np.rad2deg(spacing) try: projection = ax.projection except AttributeError: print( "The ax.projection does not exist. You must set projection to plot Cartopy maps, such as ax = plt.subplot(211, projection=cartopy.crs.PlateCarree())" ) projection = None if isinstance(projection, ccrs.PlateCarree): spacing = math.degrees(spacing) else: spacing = spacing * EARTH_RADIUS * 1e3 if aspect is None: aspect = 2.0 / 3.0 if size is None: size = spacing * 0.5 height = size * aspect trench_left_features = shapelify_feature_lines( self.trench_left, tessellate_degrees=tessellate_degrees, central_meridian=central_meridian, ) trench_right_features = shapelify_feature_lines( self.trench_right, tessellate_degrees=tessellate_degrees, central_meridian=central_meridian, ) plot_subduction_teeth( trench_left_features, size, "l", height, spacing, projection=projection, ax=ax, color=color, **kwargs, ) plot_subduction_teeth( trench_right_features, size, "r", height, spacing, projection=projection, ax=ax, color=color, **kwargs, ) def plot_plate_id(self, ax, plate_id, **kwargs): """Plot a plate polygon with an associated `plate_id` onto a standard map Projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. plate_id : int A plate ID that identifies the continental polygon to plot. See the [Global EarthByte plate IDs list](https://www.earthbyte.org/webdav/ftp/earthbyte/GPlates/SampleData/FeatureCollections/Rotations/Global_EarthByte_PlateIDs_20071218.pdf) for a full list of plate IDs to plot. **kwargs : Keyword arguments for map presentation parameters such as `alpha`, etc. for plotting the grid. See `Matplotlib`'s `imshow` keyword arguments [here](https://matplotlib.org/3.5.1/api/_as_gen/matplotlib.axes.Axes.imshow.html). """ tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) for feature in self.topologies: if feature.get_reconstruction_plate_id() == plate_id: ft_plate = shapelify_feature_polygons( [feature], central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) return ax.add_geometries(ft_plate, crs=self.base_projection, **kwargs) def plot_grid(self, ax, grid, extent=[-180, 180, -90, 90], **kwargs): """Plot a `MaskedArray` raster or grid onto a standard map Projection. Notes ----- Uses Matplotlib's `imshow` [function](https://matplotlib.org/3.5.1/api/_as_gen/matplotlib.axes.Axes.imshow.html). Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. grid : MaskedArray or `gplately.grids.Raster` A `MaskedArray` with elements that define a grid. The number of rows in the raster corresponds to the number of latitudinal coordinates, while the number of raster columns corresponds to the number of longitudinal coordinates. extent : 1d array, default=[-180,180,-90,90] A four-element array to specify the [min lon, max lon, min lat, max lat] with which to constrain the grid image. If no extents are supplied, full global extent is assumed. **kwargs : Keyword arguments for map presentation parameters such as `alpha`, etc. for plotting the grid. See `Matplotlib`'s `imshow` keyword arguments [here](https://matplotlib.org/3.5.1/api/_as_gen/matplotlib.axes.Axes.imshow.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with the grid plotted onto the chosen map projection. """ # Override matplotlib default origin ('upper') origin = kwargs.pop("origin", "lower") from .grids import Raster if isinstance(grid, Raster): # extract extent and origin extent = grid.extent origin = grid.origin data = grid.data else: data = grid return ax.imshow( data, extent=extent, transform=self.base_projection, origin=origin, **kwargs, ) def plot_grid_from_netCDF(self, ax, filename, **kwargs): """Read a raster from a netCDF file, convert it to a `MaskedArray` and plot it onto a standard map Projection. Notes ----- `plot_grid_from_netCDF` uses Matplotlib's `imshow` [function](https://matplotlib.org/3.5.1/api/_as_gen/matplotlib.axes.Axes.imshow.html). Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. filename : str Full path to a netCDF filename. **kwargs : Keyword arguments for map presentation parameters for plotting the grid. See `Matplotlib`'s `imshow` keyword arguments [here](https://matplotlib.org/3.5.1/api/_as_gen/matplotlib.axes.Axes.imshow.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with the netCDF grid plotted onto the chosen map projection. """ # Override matplotlib default origin ('upper') origin = kwargs.pop("origin", "lower") from .grids import read_netcdf_grid raster, lon_coords, lat_coords = read_netcdf_grid(filename, return_grids=True) extent = [lon_coords[0], lon_coords[-1], lat_coords[0], lat_coords[-1]] if lon_coords[0] < lat_coords[-1]: origin = "lower" else: origin = "upper" return self.plot_grid(ax, raster, extent=extent, origin=origin, **kwargs) def plot_plate_motion_vectors( self, ax, spacingX=10, spacingY=10, normalise=False, **kwargs ): """Calculate plate motion velocity vector fields at a particular geological time and plot them onto a standard map Projection. Notes ----- `plot_plate_motion_vectors` generates a MeshNode domain of point features using given spacings in the X and Y directions (`spacingX` and `spacingY`). Each point in the domain is assigned a plate ID, and these IDs are used to obtain equivalent stage rotations of identified tectonic plates over a 5 Ma time interval. Each point and its stage rotation are used to calculate plate velocities at a particular geological time. Velocities for each domain point are represented in the north-east-down coordinate system and plotted on a GeoAxes. Vector fields can be optionally normalised by setting `normalise` to `True`. This makes vector arrow lengths uniform. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. spacingX : int, default=10 The spacing in the X direction used to make the velocity domain point feature mesh. spacingY : int, default=10 The spacing in the Y direction used to make the velocity domain point feature mesh. normalise : bool, default=False Choose whether to normalise the velocity magnitudes so that vector lengths are all equal. **kwargs : Keyword arguments for quiver presentation parameters for plotting the velocity vector field. See `Matplotlib` quiver keyword arguments [here](https://matplotlib.org/3.5.1/api/_as_gen/matplotlib.axes.Axes.quiver.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with the velocity vector field plotted onto the chosen map projection. """ lons = np.arange(-180, 180 + spacingX, spacingX) lats = np.arange(-90, 90 + spacingY, spacingY) lonq, latq = np.meshgrid(lons, lats) # create a feature from all the points velocity_domain_features = ptt.velocity_tools.make_GPML_velocity_feature( lonq.ravel(), latq.ravel() ) rotation_model = self.plate_reconstruction.rotation_model topology_features = self.plate_reconstruction.topology_features delta_time = 5.0 all_velocities = ptt.velocity_tools.get_plate_velocities( velocity_domain_features, topology_features, rotation_model, self.time, delta_time, "vector_comp", ) X, Y, U, V = ptt.velocity_tools.get_x_y_u_v(lons, lats, all_velocities) if normalise: mag = np.hypot(U, V) mag[mag == 0] = 1 U /= mag V /= mag with warnings.catch_warnings(): warnings.simplefilter("ignore", UserWarning) quiver = ax.quiver(X, Y, U, V, transform=self.base_projection, **kwargs) return quiver def get_continental_rifts( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed contiental rift lines. Notes ----- The `continental_rifts` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `continental_rifts` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `continental_rifts` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `continental_rifts` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No continental rifts have been resolved. Set `PlotTopologies.time` to construct them." ) if self.continental_rifts is None: raise ValueError("No continental rifts passed to PlotTopologies.") continental_rift_lines = shapelify_feature_lines( self.continental_rifts, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame( {"geometry": continental_rift_lines}, geometry="geometry" ) return gdf def plot_continental_rifts(self, ax, color="black", **kwargs): """Plot continental rifts on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with continental rifts plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_continental_rifts( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def get_faults(self, central_meridian=0.0, tessellate_degrees=None): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed fault lines. Notes ----- The `faults` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `faults` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `faults` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `faults` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No faults have been resolved. Set `PlotTopologies.time` to construct them." ) if self.faults is None: raise ValueError("No faults passed to PlotTopologies.") fault_lines = shapelify_feature_lines( self.faults, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": fault_lines}, geometry="geometry") return gdf def plot_faults(self, ax, color="black", **kwargs): """Plot faults on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with faults plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_faults( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def get_fracture_zones(self, central_meridian=0.0, tessellate_degrees=None): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed fracture zone lines. Notes ----- The `fracture_zones` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `fracture_zones` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `fracture_zones` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `fracture_zones` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No fracture zones have been resolved. Set `PlotTopologies.time` to construct them." ) if self.fracture_zones is None: raise ValueError("No fracture zones passed to PlotTopologies.") fracture_zone_lines = shapelify_feature_lines( self.fracture_zones, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": fracture_zone_lines}, geometry="geometry") return gdf def plot_fracture_zones(self, ax, color="black", **kwargs): """Plot fracture zones on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with fracture zones plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_fracture_zones( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def get_inferred_paleo_boundaries( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed inferred paleo boundary lines. Notes ----- The `inferred_paleo_boundaries` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `inferred_paleo_boundaries` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `inferred_paleo_boundaries` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `inferred_paleo_boundaries` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No inferred paleo boundaries have been resolved. Set `PlotTopologies.time` to construct them." ) if self.inferred_paleo_boundaries is None: raise ValueError("No inferred paleo boundaries passed to PlotTopologies.") inferred_paleo_boundary_lines = shapelify_feature_lines( self.inferred_paleo_boundaries, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame( {"geometry": inferred_paleo_boundary_lines}, geometry="geometry" ) return gdf def plot_inferred_paleo_boundaries(self, ax, color="black", **kwargs): """Plot inferred paleo boundaries on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with inferred paleo boundaries plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_inferred_paleo_boundaries( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def get_terrane_boundaries( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed terrane boundary lines. Notes ----- The `terrane_boundaries` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `terrane_boundaries` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `terrane_boundaries` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `terrane_boundaries` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No terrane boundaries have been resolved. Set `PlotTopologies.time` to construct them." ) if self.terrane_boundaries is None: raise ValueError("No terrane boundaries passed to PlotTopologies.") terrane_boundary_lines = shapelify_feature_lines( self.terrane_boundaries, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame( {"geometry": terrane_boundary_lines}, geometry="geometry" ) return gdf def plot_terrane_boundaries(self, ax, color="black", **kwargs): """Plot terrane boundaries on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with terrane boundaries plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_terrane_boundaries( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def get_transitional_crusts( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed transitional crust lines. Notes ----- The `transitional_crusts` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `transitional_crusts` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `transitional_crusts` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `transitional_crusts` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No transitional crusts have been resolved. Set `PlotTopologies.time` to construct them." ) if self.transitional_crusts is None: raise ValueError("No transitional crusts passed to PlotTopologies.") transitional_crust_lines = shapelify_feature_lines( self.transitional_crusts, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame( {"geometry": transitional_crust_lines}, geometry="geometry" ) return gdf def plot_transitional_crusts(self, ax, color="black", **kwargs): """Plot transitional crust on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with transitional crust plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_transitional_crusts( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def get_orogenic_belts( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed orogenic belt lines. Notes ----- The `orogenic_belts` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `orogenic_belts` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `orogenic_belts` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `orogenic_belts` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No orogenic belts have been resolved. Set `PlotTopologies.time` to construct them." ) if self.orogenic_belts is None: raise ValueError("No orogenic belts passed to PlotTopologies.") orogenic_belt_lines = shapelify_feature_lines( self.orogenic_belts, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": orogenic_belt_lines}, geometry="geometry") return gdf def plot_orogenic_belts(self, ax, color="black", **kwargs): """Plot orogenic belts on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with orogenic belts plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_orogenic_belts( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def get_sutures(self, central_meridian=0.0, tessellate_degrees=None): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed suture lines. Notes ----- The `sutures` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `sutures` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `sutures` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `sutures` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No sutures have been resolved. Set `PlotTopologies.time` to construct them." ) if self.sutures is None: raise ValueError("No sutures passed to PlotTopologies.") suture_lines = shapelify_feature_lines( self.sutures, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": suture_lines}, geometry="geometry") return gdf def plot_sutures(self, ax, color="black", **kwargs): """Plot sutures on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with sutures plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_sutures( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def get_continental_crusts( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed continental crust lines. Notes ----- The `continental_crusts` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `continental_crusts` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `continental_crusts` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `continental_crusts` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No continental crust topologies have been resolved. Set `PlotTopologies.time` to construct them." ) if self.continental_crusts is None: raise ValueError( "No continental crust topologies passed to PlotTopologies." ) continental_crust_lines = shapelify_feature_lines( self.continental_crusts, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame( {"geometry": continental_crust_lines}, geometry="geometry" ) return gdf def plot_continental_crusts(self, ax, color="black", **kwargs): """Plot continental crust lines on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with continental crust lines plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_continental_crusts( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def get_extended_continental_crusts( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed extended continental crust lines. Notes ----- The `extended_continental_crusts` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `extended_continental_crusts` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `extended_continental_crusts` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `extended_continental_crusts` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No extended continental crust topologies have been resolved. Set `PlotTopologies.time` to construct them." ) if self.extended_continental_crusts is None: raise ValueError( "No extended continental crust topologies passed to PlotTopologies." ) extended_continental_crust_lines = shapelify_feature_lines( self.extended_continental_crusts, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame( {"geometry": extended_continental_crust_lines}, geometry="geometry" ) return gdf def plot_extended_continental_crusts(self, ax, color="black", **kwargs): """Plot extended continental crust lines on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with extended continental crust lines plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_extended_continental_crusts( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def get_passive_continental_boundaries( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed passive continental boundary lines. Notes ----- The `passive_continental_boundaries` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `passive_continental_boundaries` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `passive_continental_boundaries` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `passive_continental_boundaries` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No passive continental boundaries have been resolved. Set `PlotTopologies.time` to construct them." ) if self.passive_continental_boundaries is None: raise ValueError( "No passive continental boundaries passed to PlotTopologies." ) passive_continental_boundary_lines = shapelify_feature_lines( self.passive_continental_boundaries, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame( {"geometry": passive_continental_boundary_lines}, geometry="geometry" ) return gdf def plot_passive_continental_boundaries(self, ax, color="black", **kwargs): """Plot passive continental boundaries on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with passive continental boundaries plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_passive_continental_boundaries( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def get_slab_edges(self, central_meridian=0.0, tessellate_degrees=None): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed slab edge lines. Notes ----- The `slab_edges` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `slab_edges` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `slab_edges` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `slab_edges` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No slab edges have been resolved. Set `PlotTopologies.time` to construct them." ) if self.slab_edges is None: raise ValueError("No slab edges passed to PlotTopologies.") slab_edge_lines = shapelify_feature_lines( self.slab_edges, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": slab_edge_lines}, geometry="geometry") return gdf def plot_slab_edges(self, ax, color="black", **kwargs): """Plot slab edges on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with slab edges plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_slab_edges( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def get_misc_transforms( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed misc transform lines. Notes ----- The `misc_transforms` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `misc_transforms` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `misc_transforms` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `misc_transforms` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No miscellaneous transforms have been resolved. Set `PlotTopologies.time` to construct them." ) if self.misc_transforms is None: raise ValueError("No miscellaneous transforms passed to PlotTopologies.") misc_transform_lines = shapelify_feature_lines( self.misc_transforms, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": misc_transform_lines}, geometry="geometry") return gdf def plot_misc_transforms(self, ax, color="black", **kwargs): """Plot miscellaneous transform boundaries on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with miscellaneous transform boundaries plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_misc_transforms( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def get_unclassified_features( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed unclassified feature lines. Notes ----- The `unclassified_features` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `unclassified_features` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `unclassified_features` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `unclassified_features` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No unclassified features have been resolved. Set `PlotTopologies.time` to construct them." ) if self.unclassified_features is None: raise ValueError("No unclassified features passed to PlotTopologies.") unclassified_feature_lines = shapelify_feature_lines( self.unclassified_features, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame( {"geometry": unclassified_feature_lines}, geometry="geometry" ) return gdf def plot_unclassified_features(self, ax, color="black", **kwargs): """Plot GPML unclassified features on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with unclassified features plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_unclassified_features( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def get_all_topologies( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed unclassified feature lines. Notes ----- The `topologies` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `topologies` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `topologies` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `topologies` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No topologies have been resolved. Set `PlotTopologies.time` to construct them." ) if self.topologies is None: raise ValueError("No topologies passed to PlotTopologies.") all_topologies = shapelify_features( self.topologies, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) # get plate IDs and feature types to add to geodataframe plate_IDs = [] feature_types = [] feature_names = [] for topo in self.topologies: ft_type = topo.get_feature_type() plate_IDs.append(topo.get_reconstruction_plate_id()) feature_types.append(ft_type) feature_names.append(ft_type.get_name()) gdf = gpd.GeoDataFrame( { "geometry": all_topologies, "reconstruction_plate_ID": plate_IDs, "feature_type": feature_types, "feature_name": feature_names, }, geometry="geometry", ) return gdf def plot_all_topologies(self, ax, color="black", **kwargs): """Plot all topologies on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with unclassified features plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_all_topologies( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def get_all_topological_sections( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of resolved topological sections. Parameters ---------- central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Returns ------- geopandas.GeoDataFrame A pandas.DataFrame that has a column with `topologies` geometry. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `topologies` to the requested `time` and thus populate the GeoDataFrame. Notes ----- The `topologies` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 # Ma gplot.time = time ...after which this function can be re-run. Once the `topologies` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. """ if self._time is None: raise ValueError( "No topologies have been resolved. Set `PlotTopologies.time` to construct them." ) if self.topologies is None: raise ValueError("No topologies passed to PlotTopologies.") topologies_list = [ *self.ridge_transforms, *self.ridges, *self.transforms, *self.trenches, *self.trench_left, *self.trench_right, *self.other, ] # get plate IDs and feature types to add to geodataframe geometries = [] plate_IDs = [] feature_types = [] feature_names = [] for topo in topologies_list: converted = shapelify_features( topo, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if not isinstance(converted, BaseGeometry): if len(converted) > 1: tmp = [] for i in converted: if isinstance(i, BaseMultipartGeometry): tmp.extend(list(i.geoms)) else: tmp.append(i) converted = tmp del tmp converted = linemerge(converted) elif len(converted) == 1: converted = converted[0] else: continue geometries.append(converted) plate_IDs.append(topo.get_reconstruction_plate_id()) feature_types.append(topo.get_feature_type()) feature_names.append(topo.get_name()) gdf = gpd.GeoDataFrame( { "geometry": geometries, "reconstruction_plate_ID": plate_IDs, "feature_type": feature_types, "feature_name": feature_names, }, geometry="geometry", ) return gdf def plot_all_topological_sections(self, ax, color="black", **kwargs): """Plot all topologies on a standard map projection. Parameters ---------- ax : cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default='black' The colour of the topology lines. By default, it is set to black. **kwargs Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with unclassified features plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_all_topological_sections( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, color=color, **kwargs)Instance variables
var anchor_plate_id-
Anchor plate ID for reconstruction. Must be an integer >= 0.
Expand source code
@property def anchor_plate_id(self): """Anchor plate ID for reconstruction. Must be an integer >= 0.""" return self._anchor_plate_id var time-
The reconstruction time.
Expand source code
@property def time(self): """The reconstruction time.""" return self._time
Methods
def get_all_topological_sections(self, central_meridian=0.0, tessellate_degrees=None)-
Create a geopandas.GeoDataFrame object containing geometries of resolved topological sections.
Parameters
central_meridian:float- Central meridian around which to perform wrapping; default: 0.0.
tessellate_degrees:floatorNone- If provided, geometries will be tessellated to this resolution prior to wrapping.
Returns
geopandas.GeoDataFrame- A pandas.DataFrame that has a column with
topologiesgeometry.
Raises
ValueError- If the optional
timeparameter has not been passed toPlotTopologies. This is needed to constructtopologiesto the requestedtimeand thus populate the GeoDataFrame.
Notes
The
topologiesneeded to produce the GeoDataFrame are automatically constructed if the optionaltimeparameter is passed to thePlotTopologiesobject before calling this function.timecan be passed either whenPlotTopologiesis first called…gplot = gplately.PlotTopologies(..., time=100,...)or anytime afterwards, by setting:
time = 100 # Ma gplot.time = time…after which this function can be re-run. Once the
topologiesare reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame.Expand source code
def get_all_topological_sections( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of resolved topological sections. Parameters ---------- central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Returns ------- geopandas.GeoDataFrame A pandas.DataFrame that has a column with `topologies` geometry. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `topologies` to the requested `time` and thus populate the GeoDataFrame. Notes ----- The `topologies` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 # Ma gplot.time = time ...after which this function can be re-run. Once the `topologies` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. """ if self._time is None: raise ValueError( "No topologies have been resolved. Set `PlotTopologies.time` to construct them." ) if self.topologies is None: raise ValueError("No topologies passed to PlotTopologies.") topologies_list = [ *self.ridge_transforms, *self.ridges, *self.transforms, *self.trenches, *self.trench_left, *self.trench_right, *self.other, ] # get plate IDs and feature types to add to geodataframe geometries = [] plate_IDs = [] feature_types = [] feature_names = [] for topo in topologies_list: converted = shapelify_features( topo, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if not isinstance(converted, BaseGeometry): if len(converted) > 1: tmp = [] for i in converted: if isinstance(i, BaseMultipartGeometry): tmp.extend(list(i.geoms)) else: tmp.append(i) converted = tmp del tmp converted = linemerge(converted) elif len(converted) == 1: converted = converted[0] else: continue geometries.append(converted) plate_IDs.append(topo.get_reconstruction_plate_id()) feature_types.append(topo.get_feature_type()) feature_names.append(topo.get_name()) gdf = gpd.GeoDataFrame( { "geometry": geometries, "reconstruction_plate_ID": plate_IDs, "feature_type": feature_types, "feature_name": feature_names, }, geometry="geometry", ) return gdf def get_all_topologies(self, central_meridian=0.0, tessellate_degrees=None)-
Create a geopandas.GeoDataFrame object containing geometries of reconstructed unclassified feature lines.
Notes
The
topologiesneeded to produce the GeoDataFrame are automatically constructed if the optionaltimeparameter is passed to thePlotTopologiesobject before calling this function.timecan be passed either whenPlotTopologiesis first called…gplot = gplately.PlotTopologies(..., time=100,...)or anytime afterwards, by setting:
time = 100 #Ma gplot.time = time…after which this function can be re-run. Once the
topologiesare reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame.Returns
gdf:instanceof<geopandas.GeoDataFrame>- A pandas.DataFrame that has a column with
topologiesgeometry. central_meridian:float- Central meridian around which to perform wrapping; default: 0.0.
tessellate_degrees:floatorNone- If provided, geometries will be tessellated to this resolution prior to wrapping.
Raises
ValueError- If the optional
timeparameter has not been passed toPlotTopologies. This is needed to constructtopologiesto the requestedtimeand thus populate the GeoDataFrame.
Expand source code
def get_all_topologies( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed unclassified feature lines. Notes ----- The `topologies` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `topologies` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `topologies` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `topologies` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No topologies have been resolved. Set `PlotTopologies.time` to construct them." ) if self.topologies is None: raise ValueError("No topologies passed to PlotTopologies.") all_topologies = shapelify_features( self.topologies, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) # get plate IDs and feature types to add to geodataframe plate_IDs = [] feature_types = [] feature_names = [] for topo in self.topologies: ft_type = topo.get_feature_type() plate_IDs.append(topo.get_reconstruction_plate_id()) feature_types.append(ft_type) feature_names.append(ft_type.get_name()) gdf = gpd.GeoDataFrame( { "geometry": all_topologies, "reconstruction_plate_ID": plate_IDs, "feature_type": feature_types, "feature_name": feature_names, }, geometry="geometry", ) return gdf def get_coastlines(self, central_meridian=0.0, tessellate_degrees=None)-
Create a geopandas.GeoDataFrame object containing geometries of reconstructed coastline polygons.
Notes
The
coastlinesneeded to produce the GeoDataFrame are automatically constructed if the optionaltimeparameter is passed to thePlotTopologiesobject before calling this function.timecan be passed either whenPlotTopologiesis first called…gplot = gplately.PlotTopologies(..., time=100,...)or anytime afterwards, by setting:
time = 100 #Ma gplot.time = time…after which this function can be re-run. Once the
coastlinesare reconstructed, they are converted into Shapely polygons whose coordinates are passed to a geopandas GeoDataFrame.Returns
gdf:instanceof<geopandas.GeoDataFrame>- A pandas.DataFrame that has a column with
coastlinesgeometry. central_meridian:float- Central meridian around which to perform wrapping; default: 0.0.
tessellate_degrees:floatorNone- If provided, geometries will be tessellated to this resolution prior to wrapping.
Raises
ValueError- If the optional
timeparameter has not been passed toPlotTopologies. This is needed to constructcoastlinesto the requestedtimeand thus populate the GeoDataFrame.
Expand source code
def get_coastlines(self, central_meridian=0.0, tessellate_degrees=None): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed coastline polygons. Notes ----- The `coastlines` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `coastlines` are reconstructed, they are converted into Shapely polygons whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `coastlines` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `coastlines` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No coastlines have been resolved. Set `PlotTopologies.time` to construct coastlines." ) if self.coastlines is None: raise ValueError("Supply coastlines to PlotTopologies object") coastline_polygons = shapelify_feature_polygons( self.coastlines, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": coastline_polygons}, geometry="geometry") return gdf def get_continent_ocean_boundaries(self, central_meridian=0.0, tessellate_degrees=None)-
Create a geopandas.GeoDataFrame object containing geometries of reconstructed continent-ocean boundary lines.
Notes
The
COBsneeded to produce the GeoDataFrame are automatically constructed if the optionaltimeparameter is passed to thePlotTopologiesobject before calling this function.timecan be passed either whenPlotTopologiesis first called…gplot = gplately.PlotTopologies(..., time=100,...)or anytime afterwards, by setting:
time = 100 #Ma gplot.time = time…after which this function can be re-run. Once the
COBsare reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame.Returns
gdf:instanceof<geopandas.GeoDataFrame>- A pandas.DataFrame that has a column with
COBsgeometry. central_meridian:float- Central meridian around which to perform wrapping; default: 0.0.
tessellate_degrees:floatorNone- If provided, geometries will be tessellated to this resolution prior to wrapping.
Raises
ValueError- If the optional
timeparameter has not been passed toPlotTopologies. This is needed to constructCOBsto the requestedtimeand thus populate the GeoDataFrame.
Expand source code
def get_continent_ocean_boundaries( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed continent-ocean boundary lines. Notes ----- The `COBs` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `COBs` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `COBs` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `COBs` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No geometries have been resolved. Set `PlotTopologies.time` to construct topologies." ) if self.COBs is None: raise ValueError("Supply COBs to PlotTopologies object") COB_lines = shapelify_feature_lines( self.COBs, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": COB_lines}, geometry="geometry") return gdf def get_continental_crusts(self, central_meridian=0.0, tessellate_degrees=None)-
Create a geopandas.GeoDataFrame object containing geometries of reconstructed continental crust lines.
Notes
The
continental_crustsneeded to produce the GeoDataFrame are automatically constructed if the optionaltimeparameter is passed to thePlotTopologiesobject before calling this function.timecan be passed either whenPlotTopologiesis first called…gplot = gplately.PlotTopologies(..., time=100,...)or anytime afterwards, by setting:
time = 100 #Ma gplot.time = time…after which this function can be re-run. Once the
continental_crustsare reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame.Returns
gdf:instanceof<geopandas.GeoDataFrame>- A pandas.DataFrame that has a column with
continental_crustsgeometry. central_meridian:float- Central meridian around which to perform wrapping; default: 0.0.
tessellate_degrees:floatorNone- If provided, geometries will be tessellated to this resolution prior to wrapping.
Raises
ValueError- If the optional
timeparameter has not been passed toPlotTopologies. This is needed to constructcontinental_cruststo the requestedtimeand thus populate the GeoDataFrame.
Expand source code
def get_continental_crusts( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed continental crust lines. Notes ----- The `continental_crusts` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `continental_crusts` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `continental_crusts` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `continental_crusts` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No continental crust topologies have been resolved. Set `PlotTopologies.time` to construct them." ) if self.continental_crusts is None: raise ValueError( "No continental crust topologies passed to PlotTopologies." ) continental_crust_lines = shapelify_feature_lines( self.continental_crusts, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame( {"geometry": continental_crust_lines}, geometry="geometry" ) return gdf def get_continental_rifts(self, central_meridian=0.0, tessellate_degrees=None)-
Create a geopandas.GeoDataFrame object containing geometries of reconstructed contiental rift lines.
Notes
The
continental_riftsneeded to produce the GeoDataFrame are automatically constructed if the optionaltimeparameter is passed to thePlotTopologiesobject before calling this function.timecan be passed either whenPlotTopologiesis first called…gplot = gplately.PlotTopologies(..., time=100,...)or anytime afterwards, by setting:
time = 100 #Ma gplot.time = time…after which this function can be re-run. Once the
continental_riftsare reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame.Returns
gdf:instanceof<geopandas.GeoDataFrame>- A pandas.DataFrame that has a column with
continental_riftsgeometry. central_meridian:float- Central meridian around which to perform wrapping; default: 0.0.
tessellate_degrees:floatorNone- If provided, geometries will be tessellated to this resolution prior to wrapping.
Raises
ValueError- If the optional
timeparameter has not been passed toPlotTopologies. This is needed to constructcontinental_riftsto the requestedtimeand thus populate the GeoDataFrame.
Expand source code
def get_continental_rifts( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed contiental rift lines. Notes ----- The `continental_rifts` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `continental_rifts` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `continental_rifts` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `continental_rifts` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No continental rifts have been resolved. Set `PlotTopologies.time` to construct them." ) if self.continental_rifts is None: raise ValueError("No continental rifts passed to PlotTopologies.") continental_rift_lines = shapelify_feature_lines( self.continental_rifts, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame( {"geometry": continental_rift_lines}, geometry="geometry" ) return gdf def get_continents(self, central_meridian=0.0, tessellate_degrees=None)-
Create a geopandas.GeoDataFrame object containing geometries of reconstructed continental polygons.
Notes
The
continentsneeded to produce the GeoDataFrame are automatically constructed if the optionaltimeparameter is passed to thePlotTopologiesobject before calling this function.timecan be passed either whenPlotTopologiesis first called…gplot = gplately.PlotTopologies(..., time=100,...)or anytime afterwards, by setting:
time = 100 #Ma gplot.time = time…after which this function can be re-run. Once the
continentsare reconstructed, they are converted into Shapely polygons whose coordinates are passed to a geopandas GeoDataFrame.Returns
gdf:instanceof<geopandas.GeoDataFrame>- A pandas.DataFrame that has a column with
continentsgeometry. central_meridian:float- Central meridian around which to perform wrapping; default: 0.0.
tessellate_degrees:floatorNone- If provided, geometries will be tessellated to this resolution prior to wrapping.
Raises
ValueError- If the optional
timeparameter has not been passed toPlotTopologies. This is needed to constructcontinentsto the requestedtimeand thus populate the GeoDataFrame.
Expand source code
def get_continents(self, central_meridian=0.0, tessellate_degrees=None): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed continental polygons. Notes ----- The `continents` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `continents` are reconstructed, they are converted into Shapely polygons whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `continents` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `continents` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No continents have been resolved. Set `PlotTopologies.time` to construct continents." ) if self.continents is None: raise ValueError("Supply continents to PlotTopologies object") continent_polygons = shapelify_feature_polygons( self.continents, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": continent_polygons}, geometry="geometry") return gdf def get_extended_continental_crusts(self, central_meridian=0.0, tessellate_degrees=None)-
Create a geopandas.GeoDataFrame object containing geometries of reconstructed extended continental crust lines.
Notes
The
extended_continental_crustsneeded to produce the GeoDataFrame are automatically constructed if the optionaltimeparameter is passed to thePlotTopologiesobject before calling this function.timecan be passed either whenPlotTopologiesis first called…gplot = gplately.PlotTopologies(..., time=100,...)or anytime afterwards, by setting:
time = 100 #Ma gplot.time = time…after which this function can be re-run. Once the
extended_continental_crustsare reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame.Returns
gdf:instanceof<geopandas.GeoDataFrame>- A pandas.DataFrame that has a column with
extended_continental_crustsgeometry. central_meridian:float- Central meridian around which to perform wrapping; default: 0.0.
tessellate_degrees:floatorNone- If provided, geometries will be tessellated to this resolution prior to wrapping.
Raises
ValueError- If the optional
timeparameter has not been passed toPlotTopologies. This is needed to constructextended_continental_cruststo the requestedtimeand thus populate the GeoDataFrame.
Expand source code
def get_extended_continental_crusts( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed extended continental crust lines. Notes ----- The `extended_continental_crusts` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `extended_continental_crusts` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `extended_continental_crusts` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `extended_continental_crusts` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No extended continental crust topologies have been resolved. Set `PlotTopologies.time` to construct them." ) if self.extended_continental_crusts is None: raise ValueError( "No extended continental crust topologies passed to PlotTopologies." ) extended_continental_crust_lines = shapelify_feature_lines( self.extended_continental_crusts, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame( {"geometry": extended_continental_crust_lines}, geometry="geometry" ) return gdf def get_faults(self, central_meridian=0.0, tessellate_degrees=None)-
Create a geopandas.GeoDataFrame object containing geometries of reconstructed fault lines.
Notes
The
faultsneeded to produce the GeoDataFrame are automatically constructed if the optionaltimeparameter is passed to thePlotTopologiesobject before calling this function.timecan be passed either whenPlotTopologiesis first called…gplot = gplately.PlotTopologies(..., time=100,...)or anytime afterwards, by setting:
time = 100 #Ma gplot.time = time…after which this function can be re-run. Once the
faultsare reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame.Returns
gdf:instanceof<geopandas.GeoDataFrame>- A pandas.DataFrame that has a column with
faultsgeometry. central_meridian:float- Central meridian around which to perform wrapping; default: 0.0.
tessellate_degrees:floatorNone- If provided, geometries will be tessellated to this resolution prior to wrapping.
Raises
ValueError- If the optional
timeparameter has not been passed toPlotTopologies. This is needed to constructfaultsto the requestedtimeand thus populate the GeoDataFrame.
Expand source code
def get_faults(self, central_meridian=0.0, tessellate_degrees=None): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed fault lines. Notes ----- The `faults` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `faults` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `faults` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `faults` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No faults have been resolved. Set `PlotTopologies.time` to construct them." ) if self.faults is None: raise ValueError("No faults passed to PlotTopologies.") fault_lines = shapelify_feature_lines( self.faults, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": fault_lines}, geometry="geometry") return gdf def get_feature(self, feature, central_meridian=0.0, tessellate_degrees=None)-
Create a geopandas.GeoDataFrame object containing geometries of reconstructed features.
Notes
The feature needed to produce the GeoDataFrame should already be constructed to a
time. This function converts the feature into a set of Shapely geometries whose coordinates are passed to a geopandas GeoDataFrame.Parameters
feature:instanceof<pygplates.Feature>- A feature reconstructed to
time.
Returns
gdf:instanceof<geopandas.GeoDataFrame>- A pandas.DataFrame that has a column with
featuregeometries.
Expand source code
def get_feature( self, feature, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed features. Notes ----- The feature needed to produce the GeoDataFrame should already be constructed to a `time`. This function converts the feature into a set of Shapely geometries whose coordinates are passed to a geopandas GeoDataFrame. Parameters ---------- feature : instance of <pygplates.Feature> A feature reconstructed to `time`. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `feature` geometries. """ shp = shapelify_features( feature, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": shp}, geometry="geometry") return gdf def get_fracture_zones(self, central_meridian=0.0, tessellate_degrees=None)-
Create a geopandas.GeoDataFrame object containing geometries of reconstructed fracture zone lines.
Notes
The
fracture_zonesneeded to produce the GeoDataFrame are automatically constructed if the optionaltimeparameter is passed to thePlotTopologiesobject before calling this function.timecan be passed either whenPlotTopologiesis first called…gplot = gplately.PlotTopologies(..., time=100,...)or anytime afterwards, by setting:
time = 100 #Ma gplot.time = time…after which this function can be re-run. Once the
fracture_zonesare reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame.Returns
gdf:instanceof<geopandas.GeoDataFrame>- A pandas.DataFrame that has a column with
fracture_zonesgeometry. central_meridian:float- Central meridian around which to perform wrapping; default: 0.0.
tessellate_degrees:floatorNone- If provided, geometries will be tessellated to this resolution prior to wrapping.
Raises
ValueError- If the optional
timeparameter has not been passed toPlotTopologies. This is needed to constructfracture_zonesto the requestedtimeand thus populate the GeoDataFrame.
Expand source code
def get_fracture_zones(self, central_meridian=0.0, tessellate_degrees=None): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed fracture zone lines. Notes ----- The `fracture_zones` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `fracture_zones` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `fracture_zones` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `fracture_zones` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No fracture zones have been resolved. Set `PlotTopologies.time` to construct them." ) if self.fracture_zones is None: raise ValueError("No fracture zones passed to PlotTopologies.") fracture_zone_lines = shapelify_feature_lines( self.fracture_zones, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": fracture_zone_lines}, geometry="geometry") return gdf def get_inferred_paleo_boundaries(self, central_meridian=0.0, tessellate_degrees=None)-
Create a geopandas.GeoDataFrame object containing geometries of reconstructed inferred paleo boundary lines.
Notes
The
inferred_paleo_boundariesneeded to produce the GeoDataFrame are automatically constructed if the optionaltimeparameter is passed to thePlotTopologiesobject before calling this function.timecan be passed either whenPlotTopologiesis first called…gplot = gplately.PlotTopologies(..., time=100,...)or anytime afterwards, by setting:
time = 100 #Ma gplot.time = time…after which this function can be re-run. Once the
inferred_paleo_boundariesare reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame.Returns
gdf:instanceof<geopandas.GeoDataFrame>- A pandas.DataFrame that has a column with
inferred_paleo_boundariesgeometry. central_meridian:float- Central meridian around which to perform wrapping; default: 0.0.
tessellate_degrees:floatorNone- If provided, geometries will be tessellated to this resolution prior to wrapping.
Raises
ValueError- If the optional
timeparameter has not been passed toPlotTopologies. This is needed to constructinferred_paleo_boundariesto the requestedtimeand thus populate the GeoDataFrame.
Expand source code
def get_inferred_paleo_boundaries( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed inferred paleo boundary lines. Notes ----- The `inferred_paleo_boundaries` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `inferred_paleo_boundaries` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `inferred_paleo_boundaries` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `inferred_paleo_boundaries` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No inferred paleo boundaries have been resolved. Set `PlotTopologies.time` to construct them." ) if self.inferred_paleo_boundaries is None: raise ValueError("No inferred paleo boundaries passed to PlotTopologies.") inferred_paleo_boundary_lines = shapelify_feature_lines( self.inferred_paleo_boundaries, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame( {"geometry": inferred_paleo_boundary_lines}, geometry="geometry" ) return gdf def get_misc_boundaries(self, central_meridian=0.0, tessellate_degrees=None)-
Create a geopandas.GeoDataFrame object containing geometries of other reconstructed lines.
Notes
The
othergeometries needed to produce the GeoDataFrame are automatically constructed if the optionaltimeparameter is passed to thePlotTopologiesobject before calling this function.timecan be passed either whenPlotTopologiesis first called…gplot = gplately.PlotTopologies(..., time=100,...)or anytime afterwards, by setting:
time = 100 #Ma gplot.time = time…after which this function can be re-run. Once the
othergeometries are reconstructed, they are converted into Shapely features whose coordinates are passed to a geopandas GeoDataFrame.Returns
gdf:instanceof<geopandas.GeoDataFrame>- A pandas.DataFrame that has a column with
othergeometry. central_meridian:float- Central meridian around which to perform wrapping; default: 0.0.
tessellate_degrees:floatorNone- If provided, geometries will be tessellated to this resolution prior to wrapping.
Raises
ValueError- If the optional
timeparameter has not been passed toPlotTopologies. This is needed to constructothergeometries to the requestedtimeand thus populate the GeoDataFrame.
Expand source code
def get_misc_boundaries(self, central_meridian=0.0, tessellate_degrees=None): """Create a geopandas.GeoDataFrame object containing geometries of other reconstructed lines. Notes ----- The `other` geometries needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `other` geometries are reconstructed, they are converted into Shapely features whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `other` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `other` geometries to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No miscellaneous topologies have been resolved. Set `PlotTopologies.time` to construct them." ) if self.other is None: raise ValueError("No miscellaneous topologies passed to PlotTopologies.") lines = shapelify_features( self.other, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": lines}, geometry="geometry") return gdf def get_misc_transforms(self, central_meridian=0.0, tessellate_degrees=None)-
Create a geopandas.GeoDataFrame object containing geometries of reconstructed misc transform lines.
Notes
The
misc_transformsneeded to produce the GeoDataFrame are automatically constructed if the optionaltimeparameter is passed to thePlotTopologiesobject before calling this function.timecan be passed either whenPlotTopologiesis first called…gplot = gplately.PlotTopologies(..., time=100,...)or anytime afterwards, by setting:
time = 100 #Ma gplot.time = time…after which this function can be re-run. Once the
misc_transformsare reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame.Returns
gdf:instanceof<geopandas.GeoDataFrame>- A pandas.DataFrame that has a column with
misc_transformsgeometry. central_meridian:float- Central meridian around which to perform wrapping; default: 0.0.
tessellate_degrees:floatorNone- If provided, geometries will be tessellated to this resolution prior to wrapping.
Raises
ValueError- If the optional
timeparameter has not been passed toPlotTopologies. This is needed to constructmisc_transformsto the requestedtimeand thus populate the GeoDataFrame.
Expand source code
def get_misc_transforms( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed misc transform lines. Notes ----- The `misc_transforms` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `misc_transforms` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `misc_transforms` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `misc_transforms` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No miscellaneous transforms have been resolved. Set `PlotTopologies.time` to construct them." ) if self.misc_transforms is None: raise ValueError("No miscellaneous transforms passed to PlotTopologies.") misc_transform_lines = shapelify_feature_lines( self.misc_transforms, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": misc_transform_lines}, geometry="geometry") return gdf def get_orogenic_belts(self, central_meridian=0.0, tessellate_degrees=None)-
Create a geopandas.GeoDataFrame object containing geometries of reconstructed orogenic belt lines.
Notes
The
orogenic_beltsneeded to produce the GeoDataFrame are automatically constructed if the optionaltimeparameter is passed to thePlotTopologiesobject before calling this function.timecan be passed either whenPlotTopologiesis first called…gplot = gplately.PlotTopologies(..., time=100,...)or anytime afterwards, by setting:
time = 100 #Ma gplot.time = time…after which this function can be re-run. Once the
orogenic_beltsare reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame.Returns
gdf:instanceof<geopandas.GeoDataFrame>- A pandas.DataFrame that has a column with
orogenic_beltsgeometry. central_meridian:float- Central meridian around which to perform wrapping; default: 0.0.
tessellate_degrees:floatorNone- If provided, geometries will be tessellated to this resolution prior to wrapping.
Raises
ValueError- If the optional
timeparameter has not been passed toPlotTopologies. This is needed to constructorogenic_beltsto the requestedtimeand thus populate the GeoDataFrame.
Expand source code
def get_orogenic_belts( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed orogenic belt lines. Notes ----- The `orogenic_belts` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `orogenic_belts` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `orogenic_belts` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `orogenic_belts` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No orogenic belts have been resolved. Set `PlotTopologies.time` to construct them." ) if self.orogenic_belts is None: raise ValueError("No orogenic belts passed to PlotTopologies.") orogenic_belt_lines = shapelify_feature_lines( self.orogenic_belts, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": orogenic_belt_lines}, geometry="geometry") return gdf def get_passive_continental_boundaries(self, central_meridian=0.0, tessellate_degrees=None)-
Create a geopandas.GeoDataFrame object containing geometries of reconstructed passive continental boundary lines.
Notes
The
passive_continental_boundariesneeded to produce the GeoDataFrame are automatically constructed if the optionaltimeparameter is passed to thePlotTopologiesobject before calling this function.timecan be passed either whenPlotTopologiesis first called…gplot = gplately.PlotTopologies(..., time=100,...)or anytime afterwards, by setting:
time = 100 #Ma gplot.time = time…after which this function can be re-run. Once the
passive_continental_boundariesare reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame.Returns
gdf:instanceof<geopandas.GeoDataFrame>- A pandas.DataFrame that has a column with
passive_continental_boundariesgeometry. central_meridian:float- Central meridian around which to perform wrapping; default: 0.0.
tessellate_degrees:floatorNone- If provided, geometries will be tessellated to this resolution prior to wrapping.
Raises
ValueError- If the optional
timeparameter has not been passed toPlotTopologies. This is needed to constructpassive_continental_boundariesto the requestedtimeand thus populate the GeoDataFrame.
Expand source code
def get_passive_continental_boundaries( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed passive continental boundary lines. Notes ----- The `passive_continental_boundaries` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `passive_continental_boundaries` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `passive_continental_boundaries` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `passive_continental_boundaries` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No passive continental boundaries have been resolved. Set `PlotTopologies.time` to construct them." ) if self.passive_continental_boundaries is None: raise ValueError( "No passive continental boundaries passed to PlotTopologies." ) passive_continental_boundary_lines = shapelify_feature_lines( self.passive_continental_boundaries, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame( {"geometry": passive_continental_boundary_lines}, geometry="geometry" ) return gdf def get_ridges(self, central_meridian=0.0, tessellate_degrees=None)-
Create a geopandas.GeoDataFrame object containing geometries of reconstructed ridge lines.
Notes
The
ridgesneeded to produce the GeoDataFrame are automatically constructed if the optionaltimeparameter is passed to thePlotTopologiesobject before calling this function.timecan be passed either whenPlotTopologiesis first called…gplot = gplately.PlotTopologies(..., time=100,...)or anytime afterwards, by setting:
time = 100 #Ma gplot.time = time…after which this function can be re-run. Once the
ridgesare reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame.Returns
gdf:instanceof<geopandas.GeoDataFrame>- A pandas.DataFrame that has a column with
ridgesgeometry. central_meridian:float- Central meridian around which to perform wrapping; default: 0.0.
tessellate_degrees:floatorNone- If provided, geometries will be tessellated to this resolution prior to wrapping.
Raises
ValueError- If the optional
timeparameter has not been passed toPlotTopologies. This is needed to constructridgesto the requestedtimeand thus populate the GeoDataFrame.
Expand source code
def get_ridges(self, central_meridian=0.0, tessellate_degrees=None): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed ridge lines. Notes ----- The `ridges` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `ridges` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `ridges` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `ridges` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No ridges have been resolved. Set `PlotTopologies.time` to construct ridges." ) if self.ridges is None: raise ValueError("No ridge topologies passed to PlotTopologies.") ridge_lines = shapelify_feature_lines( self.ridges, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": ridge_lines}, geometry="geometry") return gdf def get_ridges_and_transforms(self, central_meridian=0.0, tessellate_degrees=None)-
Create a geopandas.GeoDataFrame object containing geometries of reconstructed ridge and transform lines.
Notes
The
ridge_transformsneeded to produce the GeoDataFrame are automatically constructed if the optionaltimeparameter is passed to thePlotTopologiesobject before calling this function.timecan be passed either whenPlotTopologiesis first called…gplot = gplately.PlotTopologies(..., time=100,...)or anytime afterwards, by setting:
time = 100 #Ma gplot.time = time…after which this function can be re-run. Once the
ridge_transformsare reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame.Returns
gdf:instanceof<geopandas.GeoDataFrame>- A pandas.DataFrame that has a column with
ridgesgeometry. central_meridian:float- Central meridian around which to perform wrapping; default: 0.0.
tessellate_degrees:floatorNone- If provided, geometries will be tessellated to this resolution prior to wrapping.
Raises
ValueError- If the optional
timeparameter has not been passed toPlotTopologies. This is needed to constructridge_transformsto the requestedtimeand thus populate the GeoDataFrame.
Expand source code
def get_ridges_and_transforms( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed ridge and transform lines. Notes ----- The `ridge_transforms` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `ridge_transforms` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `ridges` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `ridge_transforms` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No ridges and transforms have been resolved. Set `PlotTopologies.time` to construct ridges and transforms." ) if self.ridge_transforms is None: raise ValueError( "No ridge and transform topologies passed to PlotTopologies." ) ridge_transform_lines = shapelify_feature_lines( self.ridge_transforms, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": ridge_transform_lines}, geometry="geometry") return gdf def get_slab_edges(self, central_meridian=0.0, tessellate_degrees=None)-
Create a geopandas.GeoDataFrame object containing geometries of reconstructed slab edge lines.
Notes
The
slab_edgesneeded to produce the GeoDataFrame are automatically constructed if the optionaltimeparameter is passed to thePlotTopologiesobject before calling this function.timecan be passed either whenPlotTopologiesis first called…gplot = gplately.PlotTopologies(..., time=100,...)or anytime afterwards, by setting:
time = 100 #Ma gplot.time = time…after which this function can be re-run. Once the
slab_edgesare reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame.Returns
gdf:instanceof<geopandas.GeoDataFrame>- A pandas.DataFrame that has a column with
slab_edgesgeometry. central_meridian:float- Central meridian around which to perform wrapping; default: 0.0.
tessellate_degrees:floatorNone- If provided, geometries will be tessellated to this resolution prior to wrapping.
Raises
ValueError- If the optional
timeparameter has not been passed toPlotTopologies. This is needed to constructslab_edgesto the requestedtimeand thus populate the GeoDataFrame.
Expand source code
def get_slab_edges(self, central_meridian=0.0, tessellate_degrees=None): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed slab edge lines. Notes ----- The `slab_edges` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `slab_edges` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `slab_edges` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `slab_edges` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No slab edges have been resolved. Set `PlotTopologies.time` to construct them." ) if self.slab_edges is None: raise ValueError("No slab edges passed to PlotTopologies.") slab_edge_lines = shapelify_feature_lines( self.slab_edges, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": slab_edge_lines}, geometry="geometry") return gdf def get_subduction_direction(self)-
Create a geopandas.GeoDataFrame object containing geometries of trench directions.
Notes
The
trench_leftandtrench_rightgeometries needed to produce the GeoDataFrame are automatically constructed if the optionaltimeparameter is passed to thePlotTopologiesobject before calling this function.timecan be passed either whenPlotTopologiesis first called…gplot = gplately.PlotTopologies(..., time=100,...)or anytime afterwards, by setting:
time = 100 #Ma gplot.time = time…after which this function can be re-run. Once the
othergeometries are reconstructed, they are converted into Shapely features whose coordinates are passed to a geopandas GeoDataFrame.Returns
gdf_left:instanceof<geopandas.GeoDataFrame>- A pandas.DataFrame that has a column with
trench_leftgeometry. gdf_right:instanceof<geopandas.GeoDataFrame>- A pandas.DataFrame that has a column with
trench_rightgeometry.
Raises
ValueError- If the optional
timeparameter has not been passed toPlotTopologies. This is needed to constructtrench_leftortrench_rightgeometries to the requestedtimeand thus populate the GeoDataFrame.
Expand source code
def get_subduction_direction(self): """Create a geopandas.GeoDataFrame object containing geometries of trench directions. Notes ----- The `trench_left` and `trench_right` geometries needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `other` geometries are reconstructed, they are converted into Shapely features whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf_left : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `trench_left` geometry. gdf_right : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `trench_right` geometry. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `trench_left` or `trench_right` geometries to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No miscellaneous topologies have been resolved. Set `PlotTopologies.time` to construct them." ) if self.trench_left is None or self.trench_right is None: raise ValueError( "No trench_left or trench_right topologies passed to PlotTopologies." ) trench_left_features = shapelify_feature_lines(self.trench_left) trench_right_features = shapelify_feature_lines(self.trench_right) gdf_left = gpd.GeoDataFrame( {"geometry": trench_left_features}, geometry="geometry" ) gdf_right = gpd.GeoDataFrame( {"geometry": trench_right_features}, geometry="geometry" ) return gdf_left, gdf_right def get_sutures(self, central_meridian=0.0, tessellate_degrees=None)-
Create a geopandas.GeoDataFrame object containing geometries of reconstructed suture lines.
Notes
The
suturesneeded to produce the GeoDataFrame are automatically constructed if the optionaltimeparameter is passed to thePlotTopologiesobject before calling this function.timecan be passed either whenPlotTopologiesis first called…gplot = gplately.PlotTopologies(..., time=100,...)or anytime afterwards, by setting:
time = 100 #Ma gplot.time = time…after which this function can be re-run. Once the
suturesare reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame.Returns
gdf:instanceof<geopandas.GeoDataFrame>- A pandas.DataFrame that has a column with
suturesgeometry. central_meridian:float- Central meridian around which to perform wrapping; default: 0.0.
tessellate_degrees:floatorNone- If provided, geometries will be tessellated to this resolution prior to wrapping.
Raises
ValueError- If the optional
timeparameter has not been passed toPlotTopologies. This is needed to constructsuturesto the requestedtimeand thus populate the GeoDataFrame.
Expand source code
def get_sutures(self, central_meridian=0.0, tessellate_degrees=None): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed suture lines. Notes ----- The `sutures` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `sutures` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `sutures` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `sutures` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No sutures have been resolved. Set `PlotTopologies.time` to construct them." ) if self.sutures is None: raise ValueError("No sutures passed to PlotTopologies.") suture_lines = shapelify_feature_lines( self.sutures, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": suture_lines}, geometry="geometry") return gdf def get_terrane_boundaries(self, central_meridian=0.0, tessellate_degrees=None)-
Create a geopandas.GeoDataFrame object containing geometries of reconstructed terrane boundary lines.
Notes
The
terrane_boundariesneeded to produce the GeoDataFrame are automatically constructed if the optionaltimeparameter is passed to thePlotTopologiesobject before calling this function.timecan be passed either whenPlotTopologiesis first called…gplot = gplately.PlotTopologies(..., time=100,...)or anytime afterwards, by setting:
time = 100 #Ma gplot.time = time…after which this function can be re-run. Once the
terrane_boundariesare reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame.Returns
gdf:instanceof<geopandas.GeoDataFrame>- A pandas.DataFrame that has a column with
terrane_boundariesgeometry. central_meridian:float- Central meridian around which to perform wrapping; default: 0.0.
tessellate_degrees:floatorNone- If provided, geometries will be tessellated to this resolution prior to wrapping.
Raises
ValueError- If the optional
timeparameter has not been passed toPlotTopologies. This is needed to constructterrane_boundariesto the requestedtimeand thus populate the GeoDataFrame.
Expand source code
def get_terrane_boundaries( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed terrane boundary lines. Notes ----- The `terrane_boundaries` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `terrane_boundaries` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `terrane_boundaries` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `terrane_boundaries` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No terrane boundaries have been resolved. Set `PlotTopologies.time` to construct them." ) if self.terrane_boundaries is None: raise ValueError("No terrane boundaries passed to PlotTopologies.") terrane_boundary_lines = shapelify_feature_lines( self.terrane_boundaries, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame( {"geometry": terrane_boundary_lines}, geometry="geometry" ) return gdf def get_transforms(self, central_meridian=0.0, tessellate_degrees=None)-
Create a geopandas.GeoDataFrame object containing geometries of reconstructed transform lines.
Notes
The
transformsneeded to produce the GeoDataFrame are automatically constructed if the optionaltimeparameter is passed to thePlotTopologiesobject before calling this function.timecan be passed either whenPlotTopologiesis first called…gplot = gplately.PlotTopologies(..., time=100,...)or anytime afterwards, by setting:
time = 100 #Ma gplot.time = time…after which this function can be re-run. Once the
transformsare reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame.Returns
gdf:instanceof<geopandas.GeoDataFrame>- A pandas.DataFrame that has a column with
transformsgeometry. central_meridian:float- Central meridian around which to perform wrapping; default: 0.0.
tessellate_degrees:floatorNone- If provided, geometries will be tessellated to this resolution prior to wrapping.
Raises
ValueError- If the optional
timeparameter has not been passed toPlotTopologies. This is needed to constructtransformsto the requestedtimeand thus populate the GeoDataFrame.
Expand source code
def get_transforms(self, central_meridian=0.0, tessellate_degrees=None): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed transform lines. Notes ----- The `transforms` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `transforms` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `transforms` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `transforms` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No transforms have been resolved. Set `PlotTopologies.time` to construct transforms." ) if self.transforms is None: raise ValueError("No transform topologies passed to PlotTopologies.") transform_lines = shapelify_feature_lines( self.transforms, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": transform_lines}, geometry="geometry") return gdf def get_transitional_crusts(self, central_meridian=0.0, tessellate_degrees=None)-
Create a geopandas.GeoDataFrame object containing geometries of reconstructed transitional crust lines.
Notes
The
transitional_crustsneeded to produce the GeoDataFrame are automatically constructed if the optionaltimeparameter is passed to thePlotTopologiesobject before calling this function.timecan be passed either whenPlotTopologiesis first called…gplot = gplately.PlotTopologies(..., time=100,...)or anytime afterwards, by setting:
time = 100 #Ma gplot.time = time…after which this function can be re-run. Once the
transitional_crustsare reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame.Returns
gdf:instanceof<geopandas.GeoDataFrame>- A pandas.DataFrame that has a column with
transitional_crustsgeometry. central_meridian:float- Central meridian around which to perform wrapping; default: 0.0.
tessellate_degrees:floatorNone- If provided, geometries will be tessellated to this resolution prior to wrapping.
Raises
ValueError- If the optional
timeparameter has not been passed toPlotTopologies. This is needed to constructtransitional_cruststo the requestedtimeand thus populate the GeoDataFrame.
Expand source code
def get_transitional_crusts( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed transitional crust lines. Notes ----- The `transitional_crusts` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `transitional_crusts` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `transitional_crusts` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `transitional_crusts` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No transitional crusts have been resolved. Set `PlotTopologies.time` to construct them." ) if self.transitional_crusts is None: raise ValueError("No transitional crusts passed to PlotTopologies.") transitional_crust_lines = shapelify_feature_lines( self.transitional_crusts, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame( {"geometry": transitional_crust_lines}, geometry="geometry" ) return gdf def get_trenches(self, central_meridian=0.0, tessellate_degrees=None)-
Create a geopandas.GeoDataFrame object containing geometries of reconstructed trench lines.
Notes
The
trenchesneeded to produce the GeoDataFrame are automatically constructed if the optionaltimeparameter is passed to thePlotTopologiesobject before calling this function.timecan be passed either whenPlotTopologiesis first called…gplot = gplately.PlotTopologies(..., time=100,...)or anytime afterwards, by setting:
time = 100 #Ma gplot.time = time…after which this function can be re-run. Once the
trenchesare reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame.Returns
gdf:instanceof<geopandas.GeoDataFrame>- A pandas.DataFrame that has a column with
trenchesgeometry. central_meridian:float- Central meridian around which to perform wrapping; default: 0.0.
tessellate_degrees:floatorNone- If provided, geometries will be tessellated to this resolution prior to wrapping.
Raises
ValueError- If the optional
timeparameter has not been passed toPlotTopologies. This is needed to constructtrenchesto the requestedtimeand thus populate the GeoDataFrame.
Expand source code
def get_trenches(self, central_meridian=0.0, tessellate_degrees=None): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed trench lines. Notes ----- The `trenches` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `trenches` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `trenches` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `trenches` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No trenches have been resolved. Set `PlotTopologies.time` to construct trenches." ) if self.trenches is None: raise ValueError("No trenches passed to PlotTopologies.") trench_lines = shapelify_feature_lines( self.trenches, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame({"geometry": trench_lines}, geometry="geometry") return gdf def get_unclassified_features(self, central_meridian=0.0, tessellate_degrees=None)-
Create a geopandas.GeoDataFrame object containing geometries of reconstructed unclassified feature lines.
Notes
The
unclassified_featuresneeded to produce the GeoDataFrame are automatically constructed if the optionaltimeparameter is passed to thePlotTopologiesobject before calling this function.timecan be passed either whenPlotTopologiesis first called…gplot = gplately.PlotTopologies(..., time=100,...)or anytime afterwards, by setting:
time = 100 #Ma gplot.time = time…after which this function can be re-run. Once the
unclassified_featuresare reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame.Returns
gdf:instanceof<geopandas.GeoDataFrame>- A pandas.DataFrame that has a column with
unclassified_featuresgeometry. central_meridian:float- Central meridian around which to perform wrapping; default: 0.0.
tessellate_degrees:floatorNone- If provided, geometries will be tessellated to this resolution prior to wrapping.
Raises
ValueError- If the optional
timeparameter has not been passed toPlotTopologies. This is needed to constructunclassified_featuresto the requestedtimeand thus populate the GeoDataFrame.
Expand source code
def get_unclassified_features( self, central_meridian=0.0, tessellate_degrees=None, ): """Create a geopandas.GeoDataFrame object containing geometries of reconstructed unclassified feature lines. Notes ----- The `unclassified_features` needed to produce the GeoDataFrame are automatically constructed if the optional `time` parameter is passed to the `PlotTopologies` object before calling this function. `time` can be passed either when `PlotTopologies` is first called... gplot = gplately.PlotTopologies(..., time=100,...) or anytime afterwards, by setting: time = 100 #Ma gplot.time = time ...after which this function can be re-run. Once the `unclassified_features` are reconstructed, they are converted into Shapely lines whose coordinates are passed to a geopandas GeoDataFrame. Returns ------- gdf : instance of <geopandas.GeoDataFrame> A pandas.DataFrame that has a column with `unclassified_features` geometry. central_meridian : float Central meridian around which to perform wrapping; default: 0.0. tessellate_degrees : float or None If provided, geometries will be tessellated to this resolution prior to wrapping. Raises ------ ValueError If the optional `time` parameter has not been passed to `PlotTopologies`. This is needed to construct `unclassified_features` to the requested `time` and thus populate the GeoDataFrame. """ if self._time is None: raise ValueError( "No unclassified features have been resolved. Set `PlotTopologies.time` to construct them." ) if self.unclassified_features is None: raise ValueError("No unclassified features passed to PlotTopologies.") unclassified_feature_lines = shapelify_feature_lines( self.unclassified_features, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) gdf = gpd.GeoDataFrame( {"geometry": unclassified_feature_lines}, geometry="geometry" ) return gdf def plot_all_topological_sections(self, ax, color='black', **kwargs)-
Plot all topologies on a standard map projection.
Parameters
ax:cartopy.mpl.geoaxes.GeoAxesorcartopy.mpl.geoaxes.GeoAxesSubplot- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection. color:str, default='black'- The colour of the topology lines. By default, it is set to black.
**kwargs- Keyword arguments for parameters such as
alpha, etc. for plotting trench geometries. SeeMatplotlibkeyword arguments here.
Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with unclassified features plotted onto the chosen map projection.
Expand source code
def plot_all_topological_sections(self, ax, color="black", **kwargs): """Plot all topologies on a standard map projection. Parameters ---------- ax : cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default='black' The colour of the topology lines. By default, it is set to black. **kwargs Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with unclassified features plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_all_topological_sections( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, color=color, **kwargs) def plot_all_topologies(self, ax, color='black', **kwargs)-
Plot all topologies on a standard map projection.
Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection. color:str, default=’black’- The colour of the trench lines. By default, it is set to black.
**kwargs : Keyword arguments for parameters such as
alpha, etc. for plotting trench geometries. SeeMatplotlibkeyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with unclassified features plotted onto the chosen map projection.
Expand source code
def plot_all_topologies(self, ax, color="black", **kwargs): """Plot all topologies on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with unclassified features plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_all_topologies( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def plot_coastlines(self, ax, **kwargs)-
Plot reconstructed coastline polygons onto a standard map Projection.
Notes
The
coastlinesfor plotting are accessed from thePlotTopologiesobject'scoastlinesattribute. Thesecoastlinesare reconstructed to thetimepassed to thePlotTopologiesobject and converted into Shapely polylines. The reconstructedcoastlinesare added onto the GeoAxes or GeoAxesSubplot mapaxusing GeoPandas. Map resentation details (e.g. facecolor, edgecolor, alpha…) are permitted as keyword arguments.Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection.
**kwargs : Keyword arguments for parameters such as
facecolor,alpha, etc. for plotting coastline geometries. SeeMatplotlibkeyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with coastline features plotted onto the chosen map projection.
Expand source code
def plot_coastlines(self, ax, **kwargs): """Plot reconstructed coastline polygons onto a standard map Projection. Notes ----- The `coastlines` for plotting are accessed from the `PlotTopologies` object's `coastlines` attribute. These `coastlines` are reconstructed to the `time` passed to the `PlotTopologies` object and converted into Shapely polylines. The reconstructed `coastlines` are added onto the GeoAxes or GeoAxesSubplot map `ax` using GeoPandas. Map resentation details (e.g. facecolor, edgecolor, alpha…) are permitted as keyword arguments. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. **kwargs : Keyword arguments for parameters such as `facecolor`, `alpha`, etc. for plotting coastline geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with coastline features plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_coastlines( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, **kwargs) def plot_continent_ocean_boundaries(self, ax, **kwargs)-
Plot reconstructed continent-ocean boundary (COB) polygons onto a standard map Projection.
Notes
The
COBsfor plotting are accessed from thePlotTopologiesobject'sCOBsattribute. TheseCOBsare reconstructed to thetimepassed to thePlotTopologiesobject and converted into Shapely polylines. The reconstructedCOBsare plotted onto the GeoAxes or GeoAxesSubplot mapaxusing GeoPandas. Map presentation details (e.g.facecolor,edgecolor,alpha…) are permitted as keyword arguments.These COBs are transformed into shapely geometries and added onto the chosen map for a specific geological time (supplied to the PlotTopologies object). Map presentation details (e.g. facecolor, edgecolor, alpha…) are permitted.
Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection.
**kwargs : Keyword arguments for parameters such as
facecolor,alpha, etc. for plotting COB geometries. SeeMatplotlibkeyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with COB features plotted onto the chosen map projection.
Expand source code
def plot_continent_ocean_boundaries(self, ax, **kwargs): """Plot reconstructed continent-ocean boundary (COB) polygons onto a standard map Projection. Notes ----- The `COBs` for plotting are accessed from the `PlotTopologies` object's `COBs` attribute. These `COBs` are reconstructed to the `time` passed to the `PlotTopologies` object and converted into Shapely polylines. The reconstructed `COBs` are plotted onto the GeoAxes or GeoAxesSubplot map `ax` using GeoPandas. Map presentation details (e.g. `facecolor`, `edgecolor`, `alpha`…) are permitted as keyword arguments. These COBs are transformed into shapely geometries and added onto the chosen map for a specific geological time (supplied to the PlotTopologies object). Map presentation details (e.g. facecolor, edgecolor, alpha…) are permitted. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. **kwargs : Keyword arguments for parameters such as `facecolor`, `alpha`, etc. for plotting COB geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with COB features plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_continent_ocean_boundaries( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, **kwargs) def plot_continental_crusts(self, ax, color='black', **kwargs)-
Plot continental crust lines on a standard map projection.
Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection. color:str, default=’black’- The colour of the trench lines. By default, it is set to black.
**kwargs : Keyword arguments for parameters such as
alpha, etc. for plotting trench geometries. SeeMatplotlibkeyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with continental crust lines plotted onto the chosen map projection.
Expand source code
def plot_continental_crusts(self, ax, color="black", **kwargs): """Plot continental crust lines on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with continental crust lines plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_continental_crusts( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def plot_continental_rifts(self, ax, color='black', **kwargs)-
Plot continental rifts on a standard map projection.
Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection. color:str, default=’black’- The colour of the trench lines. By default, it is set to black.
**kwargs : Keyword arguments for parameters such as
alpha, etc. for plotting trench geometries. SeeMatplotlibkeyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with continental rifts plotted onto the chosen map projection.
Expand source code
def plot_continental_rifts(self, ax, color="black", **kwargs): """Plot continental rifts on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with continental rifts plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_continental_rifts( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def plot_continents(self, ax, **kwargs)-
Plot reconstructed continental polygons onto a standard map Projection.
Notes
The
continentsfor plotting are accessed from thePlotTopologiesobject'scontinentsattribute. Thesecontinentsare reconstructed to thetimepassed to thePlotTopologiesobject and converted into Shapely polygons. The reconstructedcoastlinesare plotted onto the GeoAxes or GeoAxesSubplot mapaxusing GeoPandas. Map presentation details (e.g. facecolor, edgecolor, alpha…) are permitted as keyword arguments.Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection.
**kwargs : Keyword arguments for parameters such as
facecolor,alpha, etc. for plotting continental geometries. SeeMatplotlibkeyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with continent features plotted onto the chosen map projection.
Expand source code
def plot_continents(self, ax, **kwargs): """Plot reconstructed continental polygons onto a standard map Projection. Notes ----- The `continents` for plotting are accessed from the `PlotTopologies` object's `continents` attribute. These `continents` are reconstructed to the `time` passed to the `PlotTopologies` object and converted into Shapely polygons. The reconstructed `coastlines` are plotted onto the GeoAxes or GeoAxesSubplot map `ax` using GeoPandas. Map presentation details (e.g. facecolor, edgecolor, alpha…) are permitted as keyword arguments. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. **kwargs : Keyword arguments for parameters such as `facecolor`, `alpha`, etc. for plotting continental geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with continent features plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_continents( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, **kwargs) def plot_extended_continental_crusts(self, ax, color='black', **kwargs)-
Plot extended continental crust lines on a standard map projection.
Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection. color:str, default=’black’- The colour of the trench lines. By default, it is set to black.
**kwargs : Keyword arguments for parameters such as
alpha, etc. for plotting trench geometries. SeeMatplotlibkeyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with extended continental crust lines plotted onto the chosen map projection.
Expand source code
def plot_extended_continental_crusts(self, ax, color="black", **kwargs): """Plot extended continental crust lines on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with extended continental crust lines plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_extended_continental_crusts( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def plot_faults(self, ax, color='black', **kwargs)-
Plot faults on a standard map projection.
Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection. color:str, default=’black’- The colour of the trench lines. By default, it is set to black.
**kwargs : Keyword arguments for parameters such as
alpha, etc. for plotting trench geometries. SeeMatplotlibkeyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with faults plotted onto the chosen map projection.
Expand source code
def plot_faults(self, ax, color="black", **kwargs): """Plot faults on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with faults plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_faults( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def plot_feature(self, ax, feature, **kwargs)-
Plot pygplates.FeatureCollection or pygplates.Feature onto a map.
Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection.
**kwargs : Keyword arguments for parameters such as
facecolor,alpha, etc. for plotting coastline geometries. SeeMatplotlibkeyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with coastline features plotted onto the chosen map projection.
Expand source code
def plot_feature(self, ax, feature, **kwargs): """Plot pygplates.FeatureCollection or pygplates.Feature onto a map. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. **kwargs : Keyword arguments for parameters such as `facecolor`, `alpha`, etc. for plotting coastline geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with coastline features plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_feature( feature, central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, **kwargs) def plot_fracture_zones(self, ax, color='black', **kwargs)-
Plot fracture zones on a standard map projection.
Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection. color:str, default=’black’- The colour of the trench lines. By default, it is set to black.
**kwargs : Keyword arguments for parameters such as
alpha, etc. for plotting trench geometries. SeeMatplotlibkeyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with fracture zones plotted onto the chosen map projection.
Expand source code
def plot_fracture_zones(self, ax, color="black", **kwargs): """Plot fracture zones on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with fracture zones plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_fracture_zones( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def plot_grid(self, ax, grid, extent=[-180, 180, -90, 90], **kwargs)-
Plot a
MaskedArrayraster or grid onto a standard map Projection.Notes
Uses Matplotlib's
imshowfunction.Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection. grid:MaskedArrayorRaster- A
MaskedArraywith elements that define a grid. The number of rows in the raster corresponds to the number of latitudinal coordinates, while the number of raster columns corresponds to the number of longitudinal coordinates. extent:1d array, default=[-180,180,-90,90]- A four-element array to specify the [min lon, max lon, min lat, max lat] with which to constrain the grid image. If no extents are supplied, full global extent is assumed.
**kwargs : Keyword arguments for map presentation parameters such as
alpha, etc. for plotting the grid. SeeMatplotlib'simshowkeyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with the grid plotted onto the chosen map projection.
Expand source code
def plot_grid(self, ax, grid, extent=[-180, 180, -90, 90], **kwargs): """Plot a `MaskedArray` raster or grid onto a standard map Projection. Notes ----- Uses Matplotlib's `imshow` [function](https://matplotlib.org/3.5.1/api/_as_gen/matplotlib.axes.Axes.imshow.html). Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. grid : MaskedArray or `gplately.grids.Raster` A `MaskedArray` with elements that define a grid. The number of rows in the raster corresponds to the number of latitudinal coordinates, while the number of raster columns corresponds to the number of longitudinal coordinates. extent : 1d array, default=[-180,180,-90,90] A four-element array to specify the [min lon, max lon, min lat, max lat] with which to constrain the grid image. If no extents are supplied, full global extent is assumed. **kwargs : Keyword arguments for map presentation parameters such as `alpha`, etc. for plotting the grid. See `Matplotlib`'s `imshow` keyword arguments [here](https://matplotlib.org/3.5.1/api/_as_gen/matplotlib.axes.Axes.imshow.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with the grid plotted onto the chosen map projection. """ # Override matplotlib default origin ('upper') origin = kwargs.pop("origin", "lower") from .grids import Raster if isinstance(grid, Raster): # extract extent and origin extent = grid.extent origin = grid.origin data = grid.data else: data = grid return ax.imshow( data, extent=extent, transform=self.base_projection, origin=origin, **kwargs, ) def plot_grid_from_netCDF(self, ax, filename, **kwargs)-
Read a raster from a netCDF file, convert it to a
MaskedArrayand plot it onto a standard map Projection.Notes
plot_grid_from_netCDFuses Matplotlib'simshowfunction.Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection. filename:str- Full path to a netCDF filename.
**kwargs : Keyword arguments for map presentation parameters for plotting the grid. See
Matplotlib'simshowkeyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with the netCDF grid plotted onto the chosen map projection.
Expand source code
def plot_grid_from_netCDF(self, ax, filename, **kwargs): """Read a raster from a netCDF file, convert it to a `MaskedArray` and plot it onto a standard map Projection. Notes ----- `plot_grid_from_netCDF` uses Matplotlib's `imshow` [function](https://matplotlib.org/3.5.1/api/_as_gen/matplotlib.axes.Axes.imshow.html). Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. filename : str Full path to a netCDF filename. **kwargs : Keyword arguments for map presentation parameters for plotting the grid. See `Matplotlib`'s `imshow` keyword arguments [here](https://matplotlib.org/3.5.1/api/_as_gen/matplotlib.axes.Axes.imshow.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with the netCDF grid plotted onto the chosen map projection. """ # Override matplotlib default origin ('upper') origin = kwargs.pop("origin", "lower") from .grids import read_netcdf_grid raster, lon_coords, lat_coords = read_netcdf_grid(filename, return_grids=True) extent = [lon_coords[0], lon_coords[-1], lat_coords[0], lat_coords[-1]] if lon_coords[0] < lat_coords[-1]: origin = "lower" else: origin = "upper" return self.plot_grid(ax, raster, extent=extent, origin=origin, **kwargs) def plot_inferred_paleo_boundaries(self, ax, color='black', **kwargs)-
Plot inferred paleo boundaries on a standard map projection.
Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection. color:str, default=’black’- The colour of the trench lines. By default, it is set to black.
**kwargs : Keyword arguments for parameters such as
alpha, etc. for plotting trench geometries. SeeMatplotlibkeyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with inferred paleo boundaries plotted onto the chosen map projection.
Expand source code
def plot_inferred_paleo_boundaries(self, ax, color="black", **kwargs): """Plot inferred paleo boundaries on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with inferred paleo boundaries plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_inferred_paleo_boundaries( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def plot_misc_boundaries(self, ax, color='black', **kwargs)-
Plot reconstructed miscellaneous plate boundary polylines onto a standard map Projection.
Notes
The miscellaneous boundary sections for plotting are accessed from the
PlotTopologiesobject'sotherattribute. Theseotherboundaries are reconstructed to thetimepassed to thePlotTopologiesobject and converted into Shapely polylines. The reconstructedotherboundaries are plotted onto the GeoAxes or GeoAxesSubplot mapaxusing GeoPandas. Map presentation details (e.g.facecolor,edgecolor,alpha…) are permitted as keyword arguments.Miscellaneous boundary geometries are wrapped to the dateline using pyGPlates' DateLineWrapper by splitting a polyline into multiple polylines at the dateline. This is to avoid horizontal lines being formed between polylines at longitudes of -180 and 180 degrees. Point features near the poles (-89 & 89 degree latitude) are also clipped to ensure compatibility with Cartopy.
Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection. color:str, default=’black’- The colour of the boundary lines. By default, it is set to black.
**kwargs : Keyword arguments for parameters such as ‘alpha’, etc. for plotting miscellaneous boundary geometries. See
Matplotlibkeyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with miscellaneous boundary features plotted onto the chosen map projection.
Expand source code
def plot_misc_boundaries(self, ax, color="black", **kwargs): """Plot reconstructed miscellaneous plate boundary polylines onto a standard map Projection. Notes ----- The miscellaneous boundary sections for plotting are accessed from the `PlotTopologies` object's `other` attribute. These `other` boundaries are reconstructed to the `time` passed to the `PlotTopologies` object and converted into Shapely polylines. The reconstructed `other` boundaries are plotted onto the GeoAxes or GeoAxesSubplot map `ax` using GeoPandas. Map presentation details (e.g. `facecolor`, `edgecolor`, `alpha`…) are permitted as keyword arguments. Miscellaneous boundary geometries are wrapped to the dateline using pyGPlates' [DateLineWrapper](https://www.gplates.org/docs/pygplates/generated/pygplates.datelinewrapper) by splitting a polyline into multiple polylines at the dateline. This is to avoid horizontal lines being formed between polylines at longitudes of -180 and 180 degrees. Point features near the poles (-89 & 89 degree latitude) are also clipped to ensure compatibility with Cartopy. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the boundary lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as ‘alpha’, etc. for plotting miscellaneous boundary geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with miscellaneous boundary features plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_misc_boundaries( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def plot_misc_transforms(self, ax, color='black', **kwargs)-
Plot miscellaneous transform boundaries on a standard map projection.
Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection. color:str, default=’black’- The colour of the trench lines. By default, it is set to black.
**kwargs : Keyword arguments for parameters such as
alpha, etc. for plotting trench geometries. SeeMatplotlibkeyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with miscellaneous transform boundaries plotted onto the chosen map projection.
Expand source code
def plot_misc_transforms(self, ax, color="black", **kwargs): """Plot miscellaneous transform boundaries on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with miscellaneous transform boundaries plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_misc_transforms( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def plot_orogenic_belts(self, ax, color='black', **kwargs)-
Plot orogenic belts on a standard map projection.
Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection. color:str, default=’black’- The colour of the trench lines. By default, it is set to black.
**kwargs : Keyword arguments for parameters such as
alpha, etc. for plotting trench geometries. SeeMatplotlibkeyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with orogenic belts plotted onto the chosen map projection.
Expand source code
def plot_orogenic_belts(self, ax, color="black", **kwargs): """Plot orogenic belts on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with orogenic belts plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_orogenic_belts( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def plot_passive_continental_boundaries(self, ax, color='black', **kwargs)-
Plot passive continental boundaries on a standard map projection.
Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection. color:str, default=’black’- The colour of the trench lines. By default, it is set to black.
**kwargs : Keyword arguments for parameters such as
alpha, etc. for plotting trench geometries. SeeMatplotlibkeyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with passive continental boundaries plotted onto the chosen map projection.
Expand source code
def plot_passive_continental_boundaries(self, ax, color="black", **kwargs): """Plot passive continental boundaries on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with passive continental boundaries plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_passive_continental_boundaries( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def plot_plate_id(self, ax, plate_id, **kwargs)-
Plot a plate polygon with an associated
plate_idonto a standard map Projection.Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection. plate_id:int- A plate ID that identifies the continental polygon to plot. See the Global EarthByte plate IDs list for a full list of plate IDs to plot.
**kwargs : Keyword arguments for map presentation parameters such as
alpha, etc. for plotting the grid. SeeMatplotlib'simshowkeyword arguments here.Expand source code
def plot_plate_id(self, ax, plate_id, **kwargs): """Plot a plate polygon with an associated `plate_id` onto a standard map Projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. plate_id : int A plate ID that identifies the continental polygon to plot. See the [Global EarthByte plate IDs list](https://www.earthbyte.org/webdav/ftp/earthbyte/GPlates/SampleData/FeatureCollections/Rotations/Global_EarthByte_PlateIDs_20071218.pdf) for a full list of plate IDs to plot. **kwargs : Keyword arguments for map presentation parameters such as `alpha`, etc. for plotting the grid. See `Matplotlib`'s `imshow` keyword arguments [here](https://matplotlib.org/3.5.1/api/_as_gen/matplotlib.axes.Axes.imshow.html). """ tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) for feature in self.topologies: if feature.get_reconstruction_plate_id() == plate_id: ft_plate = shapelify_feature_polygons( [feature], central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) return ax.add_geometries(ft_plate, crs=self.base_projection, **kwargs) def plot_plate_motion_vectors(self, ax, spacingX=10, spacingY=10, normalise=False, **kwargs)-
Calculate plate motion velocity vector fields at a particular geological time and plot them onto a standard map Projection.
Notes
plot_plate_motion_vectorsgenerates a MeshNode domain of point features using given spacings in the X and Y directions (spacingXandspacingY). Each point in the domain is assigned a plate ID, and these IDs are used to obtain equivalent stage rotations of identified tectonic plates over a 5 Ma time interval. Each point and its stage rotation are used to calculate plate velocities at a particular geological time. Velocities for each domain point are represented in the north-east-down coordinate system and plotted on a GeoAxes.Vector fields can be optionally normalised by setting
normalisetoTrue. This makes vector arrow lengths uniform.Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection. spacingX:int, default=10- The spacing in the X direction used to make the velocity domain point feature mesh.
spacingY:int, default=10- The spacing in the Y direction used to make the velocity domain point feature mesh.
normalise:bool, default=False- Choose whether to normalise the velocity magnitudes so that vector lengths are all equal.
**kwargs : Keyword arguments for quiver presentation parameters for plotting the velocity vector field. See
Matplotlibquiver keyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with the velocity vector field plotted onto the chosen map projection.
Expand source code
def plot_plate_motion_vectors( self, ax, spacingX=10, spacingY=10, normalise=False, **kwargs ): """Calculate plate motion velocity vector fields at a particular geological time and plot them onto a standard map Projection. Notes ----- `plot_plate_motion_vectors` generates a MeshNode domain of point features using given spacings in the X and Y directions (`spacingX` and `spacingY`). Each point in the domain is assigned a plate ID, and these IDs are used to obtain equivalent stage rotations of identified tectonic plates over a 5 Ma time interval. Each point and its stage rotation are used to calculate plate velocities at a particular geological time. Velocities for each domain point are represented in the north-east-down coordinate system and plotted on a GeoAxes. Vector fields can be optionally normalised by setting `normalise` to `True`. This makes vector arrow lengths uniform. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. spacingX : int, default=10 The spacing in the X direction used to make the velocity domain point feature mesh. spacingY : int, default=10 The spacing in the Y direction used to make the velocity domain point feature mesh. normalise : bool, default=False Choose whether to normalise the velocity magnitudes so that vector lengths are all equal. **kwargs : Keyword arguments for quiver presentation parameters for plotting the velocity vector field. See `Matplotlib` quiver keyword arguments [here](https://matplotlib.org/3.5.1/api/_as_gen/matplotlib.axes.Axes.quiver.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with the velocity vector field plotted onto the chosen map projection. """ lons = np.arange(-180, 180 + spacingX, spacingX) lats = np.arange(-90, 90 + spacingY, spacingY) lonq, latq = np.meshgrid(lons, lats) # create a feature from all the points velocity_domain_features = ptt.velocity_tools.make_GPML_velocity_feature( lonq.ravel(), latq.ravel() ) rotation_model = self.plate_reconstruction.rotation_model topology_features = self.plate_reconstruction.topology_features delta_time = 5.0 all_velocities = ptt.velocity_tools.get_plate_velocities( velocity_domain_features, topology_features, rotation_model, self.time, delta_time, "vector_comp", ) X, Y, U, V = ptt.velocity_tools.get_x_y_u_v(lons, lats, all_velocities) if normalise: mag = np.hypot(U, V) mag[mag == 0] = 1 U /= mag V /= mag with warnings.catch_warnings(): warnings.simplefilter("ignore", UserWarning) quiver = ax.quiver(X, Y, U, V, transform=self.base_projection, **kwargs) return quiver def plot_ridges(self, ax, color='black', **kwargs)-
Plot reconstructed ridge polylines onto a standard map Projection.
Notes
The
ridgesfor plotting are accessed from thePlotTopologiesobject'sridgesattribute. Theseridgesare reconstructed to thetimepassed to thePlotTopologiesobject and converted into Shapely polylines. The reconstructedridgesare plotted onto the GeoAxes or GeoAxesSubplot mapaxusing GeoPandas. Map presentation details (e.g.facecolor,edgecolor,alpha…) are permitted as keyword arguments.Ridge geometries are wrapped to the dateline using pyGPlates' DateLineWrapper by splitting a polyline into multiple polylines at the dateline. This is to avoid horizontal lines being formed between polylines at longitudes of -180 and 180 degrees. Point features near the poles (-89 & 89 degree latitude) are also clipped to ensure compatibility with Cartopy.
Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection. color:str, default=’black’- The colour of the ridge lines. By default, it is set to black.
**kwargs : Keyword arguments for parameters such as
alpha, etc. for plotting ridge geometries. SeeMatplotlibkeyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with ridge features plotted onto the chosen map projection.
Expand source code
def plot_ridges(self, ax, color="black", **kwargs): """Plot reconstructed ridge polylines onto a standard map Projection. Notes ----- The `ridges` for plotting are accessed from the `PlotTopologies` object's `ridges` attribute. These `ridges` are reconstructed to the `time` passed to the `PlotTopologies` object and converted into Shapely polylines. The reconstructed `ridges` are plotted onto the GeoAxes or GeoAxesSubplot map `ax` using GeoPandas. Map presentation details (e.g. `facecolor`, `edgecolor`, `alpha`…) are permitted as keyword arguments. Ridge geometries are wrapped to the dateline using pyGPlates' [DateLineWrapper](https://www.gplates.org/docs/pygplates/generated/pygplates.datelinewrapper) by splitting a polyline into multiple polylines at the dateline. This is to avoid horizontal lines being formed between polylines at longitudes of -180 and 180 degrees. Point features near the poles (-89 & 89 degree latitude) are also clipped to ensure compatibility with Cartopy. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the ridge lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting ridge geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with ridge features plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_ridges( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def plot_ridges_and_transforms(self, ax, color='black', **kwargs)-
Plot reconstructed ridge & transform boundary polylines onto a standard map Projection.
Notes
The ridge & transform sections for plotting are accessed from the
PlotTopologiesobject'sridge_transformsattribute. Theseridge_transformsare reconstructed to thetimepassed to thePlotTopologiesobject and converted into Shapely polylines. The reconstructedridge_transformsare plotted onto the GeoAxes or GeoAxesSubplot mapaxusing GeoPandas. Map presentation details (e.g.facecolor,edgecolor,alpha…) are permitted as keyword arguments.Note: Ridge & transform geometries are wrapped to the dateline using pyGPlates' DateLineWrapper by splitting a polyline into multiple polylines at the dateline. This is to avoid horizontal lines being formed between polylines at longitudes of -180 and 180 degrees. Point features near the poles (-89 & 89 degree latitude) are also clipped to ensure compatibility with Cartopy.
Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection. color:str, default=’black’- The colour of the ridge & transform lines. By default, it is set to black.
**kwargs : Keyword arguments for parameters such as ‘alpha’, etc. for plotting ridge & transform geometries. See
Matplotlibkeyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with ridge & transform features plotted onto the chosen map projection.
Expand source code
def plot_ridges_and_transforms(self, ax, color="black", **kwargs): """Plot reconstructed ridge & transform boundary polylines onto a standard map Projection. Notes ----- The ridge & transform sections for plotting are accessed from the `PlotTopologies` object's `ridge_transforms` attribute. These `ridge_transforms` are reconstructed to the `time` passed to the `PlotTopologies` object and converted into Shapely polylines. The reconstructed `ridge_transforms` are plotted onto the GeoAxes or GeoAxesSubplot map `ax` using GeoPandas. Map presentation details (e.g. `facecolor`, `edgecolor`, `alpha`…) are permitted as keyword arguments. Note: Ridge & transform geometries are wrapped to the dateline using pyGPlates' [DateLineWrapper](https://www.gplates.org/docs/pygplates/generated/pygplates.datelinewrapper) by splitting a polyline into multiple polylines at the dateline. This is to avoid horizontal lines being formed between polylines at longitudes of -180 and 180 degrees. Point features near the poles (-89 & 89 degree latitude) are also clipped to ensure compatibility with Cartopy. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the ridge & transform lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as ‘alpha’, etc. for plotting ridge & transform geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with ridge & transform features plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_ridges_and_transforms( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def plot_slab_edges(self, ax, color='black', **kwargs)-
Plot slab edges on a standard map projection.
Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection. color:str, default=’black’- The colour of the trench lines. By default, it is set to black.
**kwargs : Keyword arguments for parameters such as
alpha, etc. for plotting trench geometries. SeeMatplotlibkeyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with slab edges plotted onto the chosen map projection.
Expand source code
def plot_slab_edges(self, ax, color="black", **kwargs): """Plot slab edges on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with slab edges plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_slab_edges( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def plot_subduction_teeth(self, ax, spacing=0.07, size=None, aspect=None, color='black', **kwargs)-
Plot subduction teeth onto a standard map Projection.
Notes
Subduction teeth are tessellated from
PlotTopologiesobject attributestrench_leftandtrench_right, and transformed into Shapely polygons for plotting.Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection. spacing:float, default=0.07- The tessellation threshold (in radians). Parametrises subduction tooth density.
Triangles are generated only along line segments with distances that exceed
the given threshold
spacing. size:float, default=None- Length of teeth triangle base (in radians). If kept at
None, thensize = 0.5*spacing. aspect:float, default=None- Aspect ratio of teeth triangles. If kept at
None, thenaspect = 2/3*size. color:str, default='black'- The colour of the teeth. By default, it is set to black.
**kwargs : Keyword arguments parameters such as
alpha, etc. for plotting subduction tooth polygons. SeeMatplotlibkeyword arguments here.Expand source code
def plot_subduction_teeth( self, ax, spacing=0.07, size=None, aspect=None, color="black", **kwargs ): """Plot subduction teeth onto a standard map Projection. Notes ----- Subduction teeth are tessellated from `PlotTopologies` object attributes `trench_left` and `trench_right`, and transformed into Shapely polygons for plotting. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. spacing : float, default=0.07 The tessellation threshold (in radians). Parametrises subduction tooth density. Triangles are generated only along line segments with distances that exceed the given threshold `spacing`. size : float, default=None Length of teeth triangle base (in radians). If kept at `None`, then `size = 0.5*spacing`. aspect : float, default=None Aspect ratio of teeth triangles. If kept at `None`, then `aspect = 2/3*size`. color : str, default='black' The colour of the teeth. By default, it is set to black. **kwargs : Keyword arguments parameters such as `alpha`, etc. for plotting subduction tooth polygons. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). """ if self._time is None: raise ValueError( "No topologies have been resolved. Set `PlotTopologies.time` to construct them." ) if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") central_meridian = _meridian_from_ax(ax) tessellate_degrees = np.rad2deg(spacing) try: projection = ax.projection except AttributeError: print( "The ax.projection does not exist. You must set projection to plot Cartopy maps, such as ax = plt.subplot(211, projection=cartopy.crs.PlateCarree())" ) projection = None if isinstance(projection, ccrs.PlateCarree): spacing = math.degrees(spacing) else: spacing = spacing * EARTH_RADIUS * 1e3 if aspect is None: aspect = 2.0 / 3.0 if size is None: size = spacing * 0.5 height = size * aspect trench_left_features = shapelify_feature_lines( self.trench_left, tessellate_degrees=tessellate_degrees, central_meridian=central_meridian, ) trench_right_features = shapelify_feature_lines( self.trench_right, tessellate_degrees=tessellate_degrees, central_meridian=central_meridian, ) plot_subduction_teeth( trench_left_features, size, "l", height, spacing, projection=projection, ax=ax, color=color, **kwargs, ) plot_subduction_teeth( trench_right_features, size, "r", height, spacing, projection=projection, ax=ax, color=color, **kwargs, ) def plot_subduction_teeth_deprecated(self, ax, spacing=0.1, size=2.0, aspect=1, color='black', **kwargs)-
Plot subduction teeth onto a standard map Projection.
Notes
Subduction teeth are tessellated from
PlotTopologiesobject attributestrench_leftandtrench_right, and transformed into Shapely polygons for plotting.Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection. spacing:float, default=0.1- The tessellation threshold (in radians). Parametrises subduction tooth density. Triangles are generated only along line segments with distances that exceed the given threshold ‘spacing’.
size:float, default=2.0- Length of teeth triangle base.
aspect:float, default=1- Aspect ratio of teeth triangles. Ratio is 1.0 by default.
color:str, default=’black’- The colour of the teeth. By default, it is set to black.
**kwargs : Keyword arguments for parameters such as ‘alpha’, etc. for plotting subduction tooth polygons. See
Matplotlibkeyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with subduction teeth plotted onto the chosen map projection.
Expand source code
def plot_subduction_teeth_deprecated( self, ax, spacing=0.1, size=2.0, aspect=1, color="black", **kwargs ): """Plot subduction teeth onto a standard map Projection. Notes ----- Subduction teeth are tessellated from `PlotTopologies` object attributes `trench_left` and `trench_right`, and transformed into Shapely polygons for plotting. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. spacing : float, default=0.1 The tessellation threshold (in radians). Parametrises subduction tooth density. Triangles are generated only along line segments with distances that exceed the given threshold ‘spacing’. size : float, default=2.0 Length of teeth triangle base. aspect : float, default=1 Aspect ratio of teeth triangles. Ratio is 1.0 by default. color : str, default=’black’ The colour of the teeth. By default, it is set to black. **kwargs : Keyword arguments for parameters such as ‘alpha’, etc. for plotting subduction tooth polygons. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with subduction teeth plotted onto the chosen map projection. """ import shapely # add Subduction Teeth subd_xL, subd_yL = self._tessellate_triangles( self.trench_left, tesselation_radians=spacing, triangle_base_length=size, triangle_aspect=-aspect, ) subd_xR, subd_yR = self._tessellate_triangles( self.trench_right, tesselation_radians=spacing, triangle_base_length=size, triangle_aspect=aspect, ) teeth = [] for tX, tY in zip(subd_xL, subd_yL): triangle_xy_points = np.c_[tX, tY] shp = shapely.geometry.Polygon(triangle_xy_points) teeth.append(shp) for tX, tY in zip(subd_xR, subd_yR): triangle_xy_points = np.c_[tX, tY] shp = shapely.geometry.Polygon(triangle_xy_points) teeth.append(shp) return ax.add_geometries(teeth, crs=self.base_projection, color=color, **kwargs) def plot_sutures(self, ax, color='black', **kwargs)-
Plot sutures on a standard map projection.
Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection. color:str, default=’black’- The colour of the trench lines. By default, it is set to black.
**kwargs : Keyword arguments for parameters such as
alpha, etc. for plotting trench geometries. SeeMatplotlibkeyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with sutures plotted onto the chosen map projection.
Expand source code
def plot_sutures(self, ax, color="black", **kwargs): """Plot sutures on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with sutures plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_sutures( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def plot_terrane_boundaries(self, ax, color='black', **kwargs)-
Plot terrane boundaries on a standard map projection.
Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection. color:str, default=’black’- The colour of the trench lines. By default, it is set to black.
**kwargs : Keyword arguments for parameters such as
alpha, etc. for plotting trench geometries. SeeMatplotlibkeyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with terrane boundaries plotted onto the chosen map projection.
Expand source code
def plot_terrane_boundaries(self, ax, color="black", **kwargs): """Plot terrane boundaries on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with terrane boundaries plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_terrane_boundaries( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def plot_transforms(self, ax, color='black', **kwargs)-
Plot reconstructed transform boundary polylines onto a standard map.
Notes
The transform sections for plotting are accessed from the
PlotTopologiesobject'stransformsattribute. Thesetransformsare reconstructed to thetimepassed to thePlotTopologiesobject and converted into Shapely polylines. The reconstructedtransformsare plotted onto the GeoAxes or GeoAxesSubplot mapaxusing GeoPandas. Map presentation details (e.g.facecolor,edgecolor,alpha…) are permitted as keyword arguments.Transform geometries are wrapped to the dateline using pyGPlates' DateLineWrapper by splitting a polyline into multiple polylines at the dateline. This is to avoid horizontal lines being formed between polylines at longitudes of -180 and 180 degrees. Point features near the poles (-89 & 89 degree latitude) are also clipped to ensure compatibility with Cartopy.
Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection. color:str, default=’black’- The colour of the transform lines. By default, it is set to black.
**kwargs : Keyword arguments for parameters such as
alpha, etc. for plotting transform geometries. SeeMatplotlibkeyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with transform features plotted onto the chosen map projection.
Expand source code
def plot_transforms(self, ax, color="black", **kwargs): """Plot reconstructed transform boundary polylines onto a standard map. Notes ----- The transform sections for plotting are accessed from the `PlotTopologies` object's `transforms` attribute. These `transforms` are reconstructed to the `time` passed to the `PlotTopologies` object and converted into Shapely polylines. The reconstructed `transforms` are plotted onto the GeoAxes or GeoAxesSubplot map `ax` using GeoPandas. Map presentation details (e.g. `facecolor`, `edgecolor`, `alpha`…) are permitted as keyword arguments. Transform geometries are wrapped to the dateline using pyGPlates' [DateLineWrapper](https://www.gplates.org/docs/pygplates/generated/pygplates.datelinewrapper) by splitting a polyline into multiple polylines at the dateline. This is to avoid horizontal lines being formed between polylines at longitudes of -180 and 180 degrees. Point features near the poles (-89 & 89 degree latitude) are also clipped to ensure compatibility with Cartopy. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the transform lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting transform geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with transform features plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_transforms( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def plot_transitional_crusts(self, ax, color='black', **kwargs)-
Plot transitional crust on a standard map projection.
Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection. color:str, default=’black’- The colour of the trench lines. By default, it is set to black.
**kwargs : Keyword arguments for parameters such as
alpha, etc. for plotting trench geometries. SeeMatplotlibkeyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with transitional crust plotted onto the chosen map projection.
Expand source code
def plot_transitional_crusts(self, ax, color="black", **kwargs): """Plot transitional crust on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with transitional crust plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_transitional_crusts( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def plot_trenches(self, ax, color='black', **kwargs)-
Plot reconstructed subduction trench polylines onto a standard map Projection.
Notes
The trench sections for plotting are accessed from the
PlotTopologiesobject'strenchesattribute. Thesetrenchesare reconstructed to thetimepassed to thePlotTopologiesobject and converted into Shapely polylines. The reconstructedtrenchesare plotted onto the GeoAxes or GeoAxesSubplot mapaxusing GeoPandas. Map presentation details (e.g.facecolor,edgecolor,alpha…) are permitted as keyword arguments.Trench geometries are wrapped to the dateline using pyGPlates' DateLineWrapper by splitting a polyline into multiple polylines at the dateline. This is to avoid horizontal lines being formed between polylines at longitudes of -180 and 180 degrees. Point features near the poles (-89 & 89 degree latitude) are also clipped to ensure compatibility with Cartopy.
Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection. color:str, default=’black’- The colour of the trench lines. By default, it is set to black.
**kwargs : Keyword arguments for parameters such as
alpha, etc. for plotting trench geometries. SeeMatplotlibkeyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with transform features plotted onto the chosen map projection.
Expand source code
def plot_trenches(self, ax, color="black", **kwargs): """Plot reconstructed subduction trench polylines onto a standard map Projection. Notes ----- The trench sections for plotting are accessed from the `PlotTopologies` object's `trenches` attribute. These `trenches` are reconstructed to the `time` passed to the `PlotTopologies` object and converted into Shapely polylines. The reconstructed `trenches` are plotted onto the GeoAxes or GeoAxesSubplot map `ax` using GeoPandas. Map presentation details (e.g. `facecolor`, `edgecolor`, `alpha`…) are permitted as keyword arguments. Trench geometries are wrapped to the dateline using pyGPlates' [DateLineWrapper](https://www.gplates.org/docs/pygplates/generated/pygplates.datelinewrapper) by splitting a polyline into multiple polylines at the dateline. This is to avoid horizontal lines being formed between polylines at longitudes of -180 and 180 degrees. Point features near the poles (-89 & 89 degree latitude) are also clipped to ensure compatibility with Cartopy. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with transform features plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_trenches( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def plot_unclassified_features(self, ax, color='black', **kwargs)-
Plot GPML unclassified features on a standard map projection.
Parameters
ax:instanceof<cartopy.mpl.geoaxes.GeoAxes>or<cartopy.mpl.geoaxes.GeoAxesSubplot>- A subclass of
matplotlib.axes.Axeswhich represents a map Projection. The map should be set at a particular Cartopy projection. color:str, default=’black’- The colour of the trench lines. By default, it is set to black.
**kwargs : Keyword arguments for parameters such as
alpha, etc. for plotting trench geometries. SeeMatplotlibkeyword arguments here.Returns
ax:instanceof<geopandas.GeoDataFrame.plot>- A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with unclassified features plotted onto the chosen map projection.
Expand source code
def plot_unclassified_features(self, ax, color="black", **kwargs): """Plot GPML unclassified features on a standard map projection. Parameters ---------- ax : instance of <cartopy.mpl.geoaxes.GeoAxes> or <cartopy.mpl.geoaxes.GeoAxesSubplot> A subclass of `matplotlib.axes.Axes` which represents a map Projection. The map should be set at a particular Cartopy projection. color : str, default=’black’ The colour of the trench lines. By default, it is set to black. **kwargs : Keyword arguments for parameters such as `alpha`, etc. for plotting trench geometries. See `Matplotlib` keyword arguments [here](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.plot.html). Returns ------- ax : instance of <geopandas.GeoDataFrame.plot> A standard cartopy.mpl.geoaxes.GeoAxes or cartopy.mpl.geoaxes.GeoAxesSubplot map with unclassified features plotted onto the chosen map projection. """ if "transform" in kwargs.keys(): warnings.warn( "'transform' keyword argument is ignored by PlotTopologies", UserWarning, ) kwargs.pop("transform") tessellate_degrees = kwargs.pop("tessellate_degrees", None) central_meridian = kwargs.pop("central_meridian", None) if central_meridian is None: central_meridian = _meridian_from_ax(ax) gdf = self.get_unclassified_features( central_meridian=central_meridian, tessellate_degrees=tessellate_degrees, ) if hasattr(ax, "projection"): gdf = _clean_polygons(data=gdf, projection=ax.projection) else: kwargs["transform"] = self.base_projection return gdf.plot(ax=ax, facecolor="none", edgecolor=color, **kwargs) def update_time(self, time)-
Re-reconstruct features and topologies to the time specified by the
PlotTopologiestimeattribute whenever it or the anchor plate is updated.Notes
The following
PlotTopologiesattributes are updated whenever a reconstructiontimeattribute is set:- resolved topology features (topological plates and networks)
- ridge and transform boundary sections (resolved features)
- ridge boundary sections (resolved features)
- transform boundary sections (resolved features)
- subduction boundary sections (resolved features)
- left subduction boundary sections (resolved features)
- right subduction boundary sections (resolved features)
- other boundary sections (resolved features) that are not subduction zones or mid-ocean ridges (ridge/transform)
Moreover, coastlines, continents and COBs are reconstructed to the new specified
time.Expand source code
def update_time(self, time): """Re-reconstruct features and topologies to the time specified by the `PlotTopologies` `time` attribute whenever it or the anchor plate is updated. Notes ----- The following `PlotTopologies` attributes are updated whenever a reconstruction `time` attribute is set: - resolved topology features (topological plates and networks) - ridge and transform boundary sections (resolved features) - ridge boundary sections (resolved features) - transform boundary sections (resolved features) - subduction boundary sections (resolved features) - left subduction boundary sections (resolved features) - right subduction boundary sections (resolved features) - other boundary sections (resolved features) that are not subduction zones or mid-ocean ridges (ridge/transform) Moreover, coastlines, continents and COBs are reconstructed to the new specified `time`. """ self._time = float(time) resolved_topologies = ptt.resolve_topologies.resolve_topologies_into_features( self.plate_reconstruction.rotation_model, self.plate_reconstruction.topology_features, self.time, ) ( self.topologies, self.ridge_transforms, self.ridges, self.transforms, self.trenches, self.trench_left, self.trench_right, self.other, ) = resolved_topologies # miscellaneous boundaries self.continental_rifts = [] self.faults = [] self.fracture_zones = [] self.inferred_paleo_boundaries = [] self.terrane_boundaries = [] self.transitional_crusts = [] self.orogenic_belts = [] self.sutures = [] self.continental_crusts = [] self.extended_continental_crusts = [] self.passive_continental_boundaries = [] self.slab_edges = [] self.misc_transforms = [] self.unclassified_features = [] for topol in self.other: if topol.get_feature_type() == pygplates.FeatureType.gpml_continental_rift: self.continental_rifts.append(topol) elif topol.get_feature_type() == pygplates.FeatureType.gpml_fault: self.faults.append(topol) elif topol.get_feature_type() == pygplates.FeatureType.gpml_fracture_zone: self.fracture_zones.append(topol) elif ( topol.get_feature_type() == pygplates.FeatureType.gpml_inferred_paleo_boundary ): self.inferred_paleo_boundaries.append(topol) elif ( topol.get_feature_type() == pygplates.FeatureType.gpml_terrane_boundary ): self.terrane_boundaries.append(topol) elif ( topol.get_feature_type() == pygplates.FeatureType.gpml_transitional_crust ): self.transitional_crusts.append(topol) elif topol.get_feature_type() == pygplates.FeatureType.gpml_orogenic_belt: self.orogenic_belts.append(topol) elif topol.get_feature_type() == pygplates.FeatureType.gpml_suture: self.sutures.append(topol) elif ( topol.get_feature_type() == pygplates.FeatureType.gpml_continental_crust ): self.continental_crusts.append(topol) elif ( topol.get_feature_type() == pygplates.FeatureType.gpml_extended_continental_crust ): self.extended_continental_crusts.append(topol) elif ( topol.get_feature_type() == pygplates.FeatureType.gpml_passive_continental_boundary ): self.passive_continental_boundaries.append(topol) elif topol.get_feature_type() == pygplates.FeatureType.gpml_slab_edge: self.slab_edges.append(topol) elif topol.get_feature_type() == pygplates.FeatureType.gpml_transform: self.misc_transforms.append(topol) elif ( topol.get_feature_type() == pygplates.FeatureType.gpml_unclassified_feature ): self.unclassified_features.append(topol) # reconstruct other important polygons and lines if self._coastlines: self.coastlines = self.plate_reconstruction.reconstruct( self._coastlines, self.time, from_time=0, anchor_plate_id=self.anchor_plate_id, ) if self._continents: self.continents = self.plate_reconstruction.reconstruct( self._continents, self.time, from_time=0, anchor_plate_id=self.anchor_plate_id, ) if self._COBs: self.COBs = self.plate_reconstruction.reconstruct( self._COBs, self.time, from_time=0, anchor_plate_id=self.anchor_plate_id )
class Points (plate_reconstruction, lons, lats, time=0, plate_id=None)-
Pointscontains methods to reconstruct and work with with geological point data. For example, the locations and plate velocities of point data can be calculated at a specific geologicaltime. ThePointsobject requires thePlateReconstructionobject to work because it holds therotation_modelandstatic_polygonsneeded to classify topological plates and quantify feature rotations through time.Attributes
plate_reconstruction:object pointer- Allows for the accessibility of
PlateReconstructionobject attributes:rotation_model,topology_featuesandstatic_polygonsfor use in thePointsobject if called using “self.plate_reconstruction.X”, where X is the attribute. lons:float,or1D array- A single float, or a 1D array containing the longitudes of point data.
lats:float 1D array- A single float, or a 1D array containing the latitudes of point data.
time:float, default=0- The specific geological time (Ma) at which to reconstruct the point data. By default, it is set to the present day (0 Ma).
plate_id:int, default=None- The plate ID of a particular tectonic plate on which point data lies, if known. This is obtained in
initif not provided.
Expand source code
class Points(object): """`Points` contains methods to reconstruct and work with with geological point data. For example, the locations and plate velocities of point data can be calculated at a specific geological `time`. The `Points` object requires the `PlateReconstruction` object to work because it holds the `rotation_model` and `static_polygons` needed to classify topological plates and quantify feature rotations through time. Attributes ---------- plate_reconstruction : object pointer Allows for the accessibility of `PlateReconstruction` object attributes: `rotation_model`, `topology_featues` and `static_polygons` for use in the `Points` object if called using “self.plate_reconstruction.X”, where X is the attribute. lons : float, or 1D array A single float, or a 1D array containing the longitudes of point data. lats : float 1D array A single float, or a 1D array containing the latitudes of point data. time : float, default=0 The specific geological time (Ma) at which to reconstruct the point data. By default, it is set to the present day (0 Ma). plate_id : int, default=None The plate ID of a particular tectonic plate on which point data lies, if known. This is obtained in `init` if not provided. """ def __init__(self, plate_reconstruction, lons, lats, time=0, plate_id=None): self.lons = lons self.lats = lats self.time = time self.attributes = dict() self.plate_reconstruction = plate_reconstruction self._update(lons, lats, time, plate_id) def _update(self, lons, lats, time=0, plate_id=None): # get Cartesian coordinates self.x, self.y, self.z = _tools.lonlat2xyz(lons, lats, degrees=False) # scale by average radius of the Earth self.x *= _tools.EARTH_RADIUS self.y *= _tools.EARTH_RADIUS self.z *= _tools.EARTH_RADIUS # store concatenated arrays self.lonlat = np.c_[self.lons, self.lats] self.xyz = np.c_[self.x, self.y, self.z] rotation_model = self.plate_reconstruction.rotation_model static_polygons = self.plate_reconstruction.static_polygons features = _tools.points_to_features(lons, lats, plate_id) if plate_id is not None: plate_id = np.atleast_1d(plate_id) self.features = features else: # partition using static polygons # being careful to observe 'from time' partitioned_features = pygplates.partition_into_plates( static_polygons, rotation_model, features, reconstruction_time=time ) self.features = partitioned_features plate_id = np.empty(len(self.lons), dtype=int) for i, feature in enumerate(partitioned_features): plate_id[i] = feature.get_reconstruction_plate_id() self.plate_id = plate_id self.FeatureCollection = pygplates.FeatureCollection(self.features) @property def size(self): """Number of points""" return len(self.lons) def __getstate__(self): filenames = self.plate_reconstruction.__getstate__() # add important variables from Points object filenames["lons"] = self.lons filenames["lats"] = self.lats filenames["time"] = self.time filenames["plate_id"] = self.plate_id for key in self.attributes: filenames[key] = self.attributes[key] return filenames def __setstate__(self, state): self.plate_reconstruction = PlateReconstruction( state["rotation_model"], state["topology_features"], state["static_polygons"], ) # reinstate unpicklable items self.lons = state["lons"] self.lats = state["lats"] self.time = state["time"] self.plate_id = state["plate_id"] self.attributes = dict() self._update(self.lons, self.lats, self.time, self.plate_id) for key in state: if key not in ["lons", "lats", "time", "plate_id"]: self.attributes[key] = state[key] def copy(self): """Returns a copy of the Points object Returns ------- Points A copy of the current Points object """ gpts = Points( self.plate_reconstruction, self.lons.copy(), self.lats.copy(), self.time, self.plate_id.copy(), ) gpts.add_attributes(**self.attributes.copy()) def add_attributes(self, **kwargs): """Adds the value of a feature attribute associated with a key. Example ------- # Define latitudes and longitudes to set up a Points object pt_lons = np.array([140., 150., 160.]) pt_lats = np.array([-30., -40., -50.]) gpts = gplately.Points(model, pt_lons, pt_lats) # Add the attributes a, b and c to the points in the Points object gpts.add_attributes( a=[10,2,2], b=[2,3,3], c=[30,0,0], ) print(gpts.attributes) The output would be: {'a': [10, 2, 2], 'b': [2, 3, 3], 'c': [30, 0, 0]} Parameters ---------- **kwargs : sequence of key=item/s A single key=value pair, or a sequence of key=value pairs denoting the name and value of an attribute. Notes ----- * An **assertion** is raised if the number of points in the Points object is not equal to the number of values associated with an attribute key. For example, consider an instance of the Points object with 3 points. If the points are ascribed an attribute `temperature`, there must be one `temperature` value per point, i.e. `temperature = [20, 15, 17.5]`. """ keys = kwargs.keys() for key in kwargs: attribute = kwargs[key] # make sure attribute is the same size as self.lons if type(attribute) is int or type(attribute) is float: array = np.full(self.lons.size, attribute) attribute = array elif isinstance(attribute, np.ndarray): if attribute.size == 1: array = np.full(self.lons.size, attribute, dtype=attribute.dtype) attribute = array assert ( len(attribute) == self.lons.size ), "Size mismatch, ensure attributes have the same number of entries as Points" self.attributes[key] = attribute if any(kwargs): # add these to the FeatureCollection for f, feature in enumerate(self.FeatureCollection): for key in keys: # extract value for each row in attribute val = self.attributes[key][f] # set this attribute on the feature feature.set_shapefile_attribute(key, val) def get_geopandas_dataframe(self): """Adds a shapely point `geometry` attribute to each point in the `gplately.Points` object. pandas.DataFrame that has a column with geometry Any existing point attributes are kept. Returns ------- GeoDataFrame : instance of `geopandas.GeoDataFrame` A pandas.DataFrame with rows equal to the number of points in the `gplately.Points` object, and an additional column containing a shapely `geometry` attribute. Example ------- pt_lons = np.array([140., 150., 160.]) pt_lats = np.array([-30., -40., -50.]) gpts = gplately.Points(model, pt_lons, pt_lats) # Add sample attributes a, b and c to the points in the Points object gpts.add_attributes( a=[10,2,2], b=[2,3,3], c=[30,0,0], ) gpts.get_geopandas_dataframe() ...has the output: a b c geometry 0 10 2 30 POINT (140.00000 -30.00000) 1 2 3 0 POINT (150.00000 -40.00000) 2 2 3 0 POINT (160.00000 -50.00000) """ import geopandas as gpd from shapely import geometry # create shapely points points = [] for lon, lat in zip(self.lons, self.lats): points.append(geometry.Point(lon, lat)) attributes = self.attributes.copy() attributes["geometry"] = points return gpd.GeoDataFrame(attributes, geometry="geometry") def get_geodataframe(self): """Returns the output of `Points.get_geopandas_dataframe()`. Adds a shapely point `geometry` attribute to each point in the `gplately.Points` object. pandas.DataFrame that has a column with geometry Any existing point attributes are kept. Returns ------- GeoDataFrame : instance of `geopandas.GeoDataFrame` A pandas.DataFrame with rows equal to the number of points in the `gplately.Points` object, and an additional column containing a shapely `geometry` attribute. Example ------- pt_lons = np.array([140., 150., 160.]) pt_lats = np.array([-30., -40., -50.]) gpts = gplately.Points(model, pt_lons, pt_lats) # Add sample attributes a, b and c to the points in the Points object gpts.add_attributes( a=[10,2,2], b=[2,3,3], c=[30,0,0], ) gpts.get_geopandas_dataframe() ...has the output: a b c geometry 0 10 2 30 POINT (140.00000 -30.00000) 1 2 3 0 POINT (150.00000 -40.00000) 2 2 3 0 POINT (160.00000 -50.00000) """ return self.get_geopandas_dataframe() def reconstruct(self, time, anchor_plate_id=None, return_array=False, **kwargs): """Reconstructs regular geological features, motion paths or flowlines to a specific geological time and extracts the latitudinal and longitudinal points of these features. Note: this method accesses and uses the rotation model attribute from the PointReconstruction object, and reconstructs the feature lat-lon point attributes of the Points object. Parameters ---------- time : float The specific geological time (Ma) to reconstruct features to. anchor_plate_id : int, default=None Reconstruct features with respect to a certain anchor plate. By default, reconstructions are made with respect to the anchor_plate_ID specified in the `gplately.PlateReconstruction` object, which is a default plate ID of 0 unless otherwise specified. return_array : bool, default False Return a `numpy.ndarray`, rather than a `Points` object. **reconstruct_type : ReconstructType, default=ReconstructType.feature_geometry The specific reconstruction type to generate based on input feature geometry type. Can be provided as ReconstructType.feature_geometry to only reconstruct regular feature geometries, or ReconstructType.MotionPath to only reconstruct motion path features, or ReconstructType.Flowline to only reconstruct flowline features. Generates :class:`reconstructed feature geometries<ReconstructedFeatureGeometry>’, or :class:`reconstructed motion paths<ReconstructedMotionPath>’, or :class:`reconstructed flowlines<ReconstructedFlowline>’ respectively. **group_with_feature : bool, default=False Used to group reconstructed geometries with their features. This can be useful when a feature has more than one geometry and hence more than one reconstructed geometry. The output *reconstructed_geometries* then becomes a list of tuples where each tuple contains a :class:`feature<Feature>` and a ``list`` of reconstructed geometries. Note: this keyword argument only applies when *reconstructed_geometries* is a list because exported files are always grouped with their features. This is applicable to all ReconstructType features. **export_wrap_to_dateline : bool, default=True Wrap/clip reconstructed geometries to the dateline (currently ignored). Returns ------- rlons : list of float A 1D numpy array enclosing all reconstructed point features' longitudes. rlats : list of float A 1D numpy array enclosing all reconstructed point features' latitudes. Raises ------ NotImplementedError if the starting time for reconstruction `from_time` is not equal to 0.0 """ from_time = self.time to_time = time if not anchor_plate_id: anchor_plate_id = self.plate_reconstruction.anchor_plate_id reconstructed_features = self.plate_reconstruction.reconstruct( self.features, to_time, from_time, anchor_plate_id=anchor_plate_id, **kwargs ) rlons, rlats = _tools.extract_feature_lonlat(reconstructed_features) if return_array: return rlons, rlats else: gpts = Points( self.plate_reconstruction, rlons, rlats, time=to_time, plate_id=self.plate_id, ) gpts.add_attributes(**self.attributes.copy()) return gpts def reconstruct_to_birth_age(self, ages, anchor_plate_id=None, **kwargs): """Reconstructs point features supplied to the `Points` object from the supplied initial time (`self.time`) to a range of times. The number of supplied times must equal the number of point features supplied to the Points object. Attributes ---------- ages : array Geological times to reconstruct features to. Must have the same length as the `Points `object's `self.features` attribute (which holds all point features represented on a unit length sphere in 3D Cartesian coordinates). anchor_plate_id : int, default=None Reconstruct features with respect to a certain anchor plate. By default, reconstructions are made with respect to the anchor_plate_ID specified in the `gplately.PlateReconstruction` object, which is a default plate ID of 0 unless otherwise specified. **kwargs Additional keyword arguments for the `gplately.PlateReconstruction.reconstruct` method. Raises ------ ValueError If the number of ages and number of point features supplied to the Points object are not identical. Returns ------- rlons, rlats : float The longitude and latitude coordinate lists of all point features reconstructed to all specified ages. Examples -------- To reconstruct n seed points' locations to B Ma (for this example n=2, with (lon,lat) = (78,30) and (56,22) at time=0 Ma, and we reconstruct to B=10 Ma): # Longitude and latitude of n=2 seed points pt_lon = np.array([78., 56]) pt_lat = np.array([30., 22]) # Call the Points object! gpts = gplately.Points(model, pt_lon, pt_lat) print(gpts.features[0].get_all_geometries()) # Confirms we have features represented as points on a sphere ages = numpy.linspace(10,10, len(pt_lon)) rlons, rlats = gpts.reconstruct_to_birth_age(ages) """ from_time = self.time if not anchor_plate_id: anchor_plate_id = self.plate_reconstruction.anchor_plate_id ages = np.array(ages) if len(ages) != len(self.features): raise ValueError("Number of points and ages must be identical") unique_ages = np.unique(ages) rlons = np.zeros(ages.shape) rlats = np.zeros(ages.shape) for age in unique_ages: mask_age = ages == age reconstructed_features = self.plate_reconstruction.reconstruct( self.features, age, from_time, anchor_plate_id=anchor_plate_id, **kwargs ) lons, lats = _tools.extract_feature_lonlat(reconstructed_features) rlons[mask_age] = lons[mask_age] rlats[mask_age] = lats[mask_age] return rlons, rlats def plate_velocity(self, time, delta_time=1): """Calculates the x and y components of tectonic plate velocities at a particular geological time. This method accesses and uses the `rotation_model` attribute from the `PlateReconstruction` object and uses the `Points` object's `self.features` attribute. Feature points are extracted and assigned plate IDs. These IDs are used to obtain the equivalent stage rotations of identified tectonic plates over a time interval `delta_time`. Each feature point and its stage rotation are used to calculate the point's plate velocity at a particular geological time. Obtained velocities for each domain point are represented in the north-east-down coordinate system, and their x,y Cartesian coordinate components are extracted. Parameters ---------- time : float The specific geological time (Ma) at which to calculate plate velocities. delta_time : float, default=1.0 The time increment used for generating partitioning plate stage rotations. 1.0 Ma by default. Returns ------- all_velocities.T : 2D numpy list A transposed 2D numpy list with two rows and a number of columns equal to the number of x,y Cartesian velocity components obtained (and thus the number of feature points extracted from a supplied feature). Each list column stores one point’s x,y, velocity components along its two rows. """ time = float(time) rotation_model = self.plate_reconstruction.rotation_model all_velocities = np.empty((len(self.features), 2)) for i, feature in enumerate(self.features): geometry = feature.get_geometry() partitioning_plate_id = feature.get_reconstruction_plate_id() equivalent_stage_rotation = rotation_model.get_rotation( time, partitioning_plate_id, time + delta_time ) velocity_vectors = pygplates.calculate_velocities( [geometry], equivalent_stage_rotation, delta_time, pygplates.VelocityUnits.cms_per_yr, ) velocities = ( pygplates.LocalCartesian.convert_from_geocentric_to_north_east_down( [geometry], velocity_vectors ) ) all_velocities[i] = velocities[0].get_y(), velocities[0].get_x() return list(all_velocities.T) def motion_path(self, time_array, anchor_plate_id=0, return_rate_of_motion=False): """Create a path of points to mark the trajectory of a plate's motion through geological time. Parameters ---------- time_array : arr An array of reconstruction times at which to determine the trajectory of a point on a plate. For example: import numpy as np min_time = 30 max_time = 100 time_step = 2.5 time_array = np.arange(min_time, max_time + time_step, time_step) anchor_plate_id : int, default=0 The ID of the anchor plate. return_rate_of_motion : bool, default=False Choose whether to return the rate of plate motion through time for each Returns ------- rlons : ndarray An n-dimensional array with columns containing the longitudes of the seed points at each timestep in `time_array`. There are n columns for n seed points. rlats : ndarray An n-dimensional array with columns containing the latitudes of the seed points at each timestep in `time_array`. There are n columns for n seed points. """ time_array = np.atleast_1d(time_array) # ndarrays to fill with reconstructed points and # rates of motion (if requested) rlons = np.empty((len(time_array), len(self.lons))) rlats = np.empty((len(time_array), len(self.lons))) for i, point_feature in enumerate(self.FeatureCollection): # Create the motion path feature motion_path_feature = pygplates.Feature.create_motion_path( point_feature.get_geometry(), time_array.tolist(), valid_time=(time_array.max(), time_array.min()), # relative_plate=int(self.plate_id[i]), # reconstruction_plate_id=int(anchor_plate_id)) relative_plate=int(self.plate_id[i]), reconstruction_plate_id=int(anchor_plate_id), ) reconstructed_motion_paths = self.plate_reconstruction.reconstruct( motion_path_feature, to_time=0, # from_time=0, reconstruct_type=pygplates.ReconstructType.motion_path, anchor_plate_id=int(anchor_plate_id), ) # Turn motion paths in to lat-lon coordinates for reconstructed_motion_path in reconstructed_motion_paths: trail = reconstructed_motion_path.get_motion_path().to_lat_lon_array() lon, lat = np.flipud(trail[:, 1]), np.flipud(trail[:, 0]) rlons[:, i] = lon rlats[:, i] = lat # Obtain step-plot coordinates for rate of motion if return_rate_of_motion is True: StepTimes = np.empty(((len(time_array) - 1) * 2, len(self.lons))) StepRates = np.empty(((len(time_array) - 1) * 2, len(self.lons))) # Get timestep TimeStep = [] for j in range(len(time_array) - 1): diff = time_array[j + 1] - time_array[j] TimeStep.append(diff) # Iterate over each segment in the reconstructed motion path, get the distance travelled by the moving # plate relative to the fixed plate in each time step Dist = [] for reconstructed_motion_path in reconstructed_motion_paths: for ( segment ) in reconstructed_motion_path.get_motion_path().get_segments(): Dist.append( segment.get_arc_length() * _tools.geocentric_radius( segment.get_start_point().to_lat_lon()[0] ) / 1e3 ) # Note that the motion path coordinates come out starting with the oldest time and working forwards # So, to match our 'times' array, we flip the order Dist = np.flipud(Dist) # Get rate of motion as distance per Myr Rate = np.asarray(Dist) / TimeStep # Manipulate arrays to get a step plot StepRate = np.zeros(len(Rate) * 2) StepRate[::2] = Rate StepRate[1::2] = Rate StepTime = np.zeros(len(Rate) * 2) StepTime[::2] = time_array[:-1] StepTime[1::2] = time_array[1:] # Append the nth point's step time and step rate coordinates to the ndarray StepTimes[:, i] = StepTime StepRates[:, i] = StepRate * 0.1 # cm/yr if return_rate_of_motion is True: return ( np.squeeze(rlons), np.squeeze(rlats), np.squeeze(StepTimes), np.squeeze(StepRates), ) else: return np.squeeze(rlons), np.squeeze(rlats) def flowline( self, time_array, left_plate_ID, right_plate_ID, return_rate_of_motion=False ): """Create a path of points to track plate motion away from spreading ridges over time using half-stage rotations. Parameters ---------- lons : arr An array of longitudes of points along spreading ridges. lats : arr An array of latitudes of points along spreading ridges. time_array : arr A list of times to obtain seed point locations at. left_plate_ID : int The plate ID of the polygon to the left of the spreading ridge. right_plate_ID : int The plate ID of the polygon to the right of the spreading ridge. return_rate_of_motion : bool, default False Choose whether to return a step time and step rate array for a step-plot of flowline motion. Returns ------- left_lon : ndarray The longitudes of the __left__ flowline for n seed points. There are n columns for n seed points, and m rows for m time steps in `time_array`. left_lat : ndarray The latitudes of the __left__ flowline of n seed points. There are n columns for n seed points, and m rows for m time steps in `time_array`. right_lon : ndarray The longitudes of the __right__ flowline of n seed points. There are n columns for n seed points, and m rows for m time steps in `time_array`. right_lat : ndarray The latitudes of the __right__ flowline of n seed points. There are n columns for n seed points, and m rows for m time steps in `time_array`. Examples -------- To access the ith seed point's left and right latitudes and longitudes: for i in np.arange(0,len(seed_points)): left_flowline_longitudes = left_lon[:,i] left_flowline_latitudes = left_lat[:,i] right_flowline_longitudes = right_lon[:,i] right_flowline_latitudes = right_lat[:,i] """ model = self.plate_reconstruction return model.create_flowline( self.lons, self.lats, time_array, left_plate_ID, right_plate_ID, return_rate_of_motion, ) def _get_dataframe(self): import geopandas as gpd data = dict() data["Longitude"] = self.lons data["Latitude"] = self.lats data["Plate_ID"] = self.plate_id for key in self.attributes: data[key] = self.attributes[key] return gpd.GeoDataFrame(data) def save(self, filename): """Saves the feature collection used in the Points object under a given filename to the current directory. The file format is determined from the filename extension. Parameters ---------- filename : string Can be provided as a string including the filename and the file format needed. Returns ------- Feature collection saved under given filename to current directory. """ filename = str(filename) if filename.endswith((".csv", ".txt", ".dat")): df = self._get_dataframe() df.to_csv(filename, index=False) elif filename.endswith((".xls", ".xlsx")): df = self._get_dataframe() df.to_excel(filename, index=False) elif filename.endswith("xml"): df = self._get_dataframe() df.to_xml(filename, index=False) elif filename.endswith(".gpml") or filename.endswith(".gpmlz"): self.FeatureCollection.write(filename) else: raise ValueError( "Cannot save to specified file type. Use csv, gpml, or xls file extension." )Instance variables
var size-
Number of points
Expand source code
@property def size(self): """Number of points""" return len(self.lons)
Methods
def add_attributes(self, **kwargs)-
Adds the value of a feature attribute associated with a key.
Example
# Define latitudes and longitudes to set up a Points object pt_lons = np.array([140., 150., 160.]) pt_lats = np.array([-30., -40., -50.]) gpts = gplately.Points(model, pt_lons, pt_lats) # Add the attributes a, b and c to the points in the Points object gpts.add_attributes( a=[10,2,2], b=[2,3,3], c=[30,0,0], ) print(gpts.attributes)The output would be:
{'a': [10, 2, 2], 'b': [2, 3, 3], 'c': [30, 0, 0]}Parameters
**kwargs:sequenceofkey=item/s- A single key=value pair, or a sequence of key=value pairs denoting the name and value of an attribute.
Notes
- An assertion is raised if the number of points in the Points object is not equal
to the number of values associated with an attribute key. For example, consider an instance
of the Points object with 3 points. If the points are ascribed an attribute
temperature, there must be onetemperaturevalue per point, i.e.temperature = [20, 15, 17.5].
Expand source code
def add_attributes(self, **kwargs): """Adds the value of a feature attribute associated with a key. Example ------- # Define latitudes and longitudes to set up a Points object pt_lons = np.array([140., 150., 160.]) pt_lats = np.array([-30., -40., -50.]) gpts = gplately.Points(model, pt_lons, pt_lats) # Add the attributes a, b and c to the points in the Points object gpts.add_attributes( a=[10,2,2], b=[2,3,3], c=[30,0,0], ) print(gpts.attributes) The output would be: {'a': [10, 2, 2], 'b': [2, 3, 3], 'c': [30, 0, 0]} Parameters ---------- **kwargs : sequence of key=item/s A single key=value pair, or a sequence of key=value pairs denoting the name and value of an attribute. Notes ----- * An **assertion** is raised if the number of points in the Points object is not equal to the number of values associated with an attribute key. For example, consider an instance of the Points object with 3 points. If the points are ascribed an attribute `temperature`, there must be one `temperature` value per point, i.e. `temperature = [20, 15, 17.5]`. """ keys = kwargs.keys() for key in kwargs: attribute = kwargs[key] # make sure attribute is the same size as self.lons if type(attribute) is int or type(attribute) is float: array = np.full(self.lons.size, attribute) attribute = array elif isinstance(attribute, np.ndarray): if attribute.size == 1: array = np.full(self.lons.size, attribute, dtype=attribute.dtype) attribute = array assert ( len(attribute) == self.lons.size ), "Size mismatch, ensure attributes have the same number of entries as Points" self.attributes[key] = attribute if any(kwargs): # add these to the FeatureCollection for f, feature in enumerate(self.FeatureCollection): for key in keys: # extract value for each row in attribute val = self.attributes[key][f] # set this attribute on the feature feature.set_shapefile_attribute(key, val) def copy(self)-
Expand source code
def copy(self): """Returns a copy of the Points object Returns ------- Points A copy of the current Points object """ gpts = Points( self.plate_reconstruction, self.lons.copy(), self.lats.copy(), self.time, self.plate_id.copy(), ) gpts.add_attributes(**self.attributes.copy()) def flowline(self, time_array, left_plate_ID, right_plate_ID, return_rate_of_motion=False)-
Create a path of points to track plate motion away from spreading ridges over time using half-stage rotations.
Parameters
lons:arr- An array of longitudes of points along spreading ridges.
lats:arr- An array of latitudes of points along spreading ridges.
time_array:arr- A list of times to obtain seed point locations at.
left_plate_ID:int- The plate ID of the polygon to the left of the spreading ridge.
right_plate_ID:int- The plate ID of the polygon to the right of the spreading ridge.
return_rate_of_motion:bool, defaultFalse- Choose whether to return a step time and step rate array for a step-plot of flowline motion.
Returns
left_lon:ndarray- The longitudes of the left flowline for n seed points.
There are n columns for n seed points, and m rows
for m time steps in
time_array. left_lat:ndarray- The latitudes of the left flowline of n seed points.
There are n columns for n seed points, and m rows
for m time steps in
time_array. right_lon:ndarray- The longitudes of the right flowline of n seed points.
There are n columns for n seed points, and m rows
for m time steps in
time_array. right_lat:ndarray- The latitudes of the right flowline of n seed points.
There are n columns for n seed points, and m rows
for m time steps in
time_array.
Examples
To access the ith seed point's left and right latitudes and longitudes:
for i in np.arange(0,len(seed_points)): left_flowline_longitudes = left_lon[:,i] left_flowline_latitudes = left_lat[:,i] right_flowline_longitudes = right_lon[:,i] right_flowline_latitudes = right_lat[:,i]Expand source code
def flowline( self, time_array, left_plate_ID, right_plate_ID, return_rate_of_motion=False ): """Create a path of points to track plate motion away from spreading ridges over time using half-stage rotations. Parameters ---------- lons : arr An array of longitudes of points along spreading ridges. lats : arr An array of latitudes of points along spreading ridges. time_array : arr A list of times to obtain seed point locations at. left_plate_ID : int The plate ID of the polygon to the left of the spreading ridge. right_plate_ID : int The plate ID of the polygon to the right of the spreading ridge. return_rate_of_motion : bool, default False Choose whether to return a step time and step rate array for a step-plot of flowline motion. Returns ------- left_lon : ndarray The longitudes of the __left__ flowline for n seed points. There are n columns for n seed points, and m rows for m time steps in `time_array`. left_lat : ndarray The latitudes of the __left__ flowline of n seed points. There are n columns for n seed points, and m rows for m time steps in `time_array`. right_lon : ndarray The longitudes of the __right__ flowline of n seed points. There are n columns for n seed points, and m rows for m time steps in `time_array`. right_lat : ndarray The latitudes of the __right__ flowline of n seed points. There are n columns for n seed points, and m rows for m time steps in `time_array`. Examples -------- To access the ith seed point's left and right latitudes and longitudes: for i in np.arange(0,len(seed_points)): left_flowline_longitudes = left_lon[:,i] left_flowline_latitudes = left_lat[:,i] right_flowline_longitudes = right_lon[:,i] right_flowline_latitudes = right_lat[:,i] """ model = self.plate_reconstruction return model.create_flowline( self.lons, self.lats, time_array, left_plate_ID, right_plate_ID, return_rate_of_motion, ) def get_geodataframe(self)-
Returns the output of
Points.get_geopandas_dataframe().Adds a shapely point
gplately.geometryattribute to each point in thePointsobject. pandas.DataFrame that has a column with geometry Any existing point attributes are kept.Returns
GeoDataFrame:instanceofgeopandas.GeoDataFrame- A pandas.DataFrame with rows equal to the number of points in the
Pointsobject, and an additional column containing a shapelygplately.geometryattribute.
Example
pt_lons = np.array([140., 150., 160.]) pt_lats = np.array([-30., -40., -50.]) gpts = gplately.Points(model, pt_lons, pt_lats) # Add sample attributes a, b and c to the points in the Points object gpts.add_attributes( a=[10,2,2], b=[2,3,3], c=[30,0,0], ) gpts.get_geopandas_dataframe()…has the output:
a b c geometry 0 10 2 30 POINT (140.00000 -30.00000) 1 2 3 0 POINT (150.00000 -40.00000) 2 2 3 0 POINT (160.00000 -50.00000)Expand source code
def get_geodataframe(self): """Returns the output of `Points.get_geopandas_dataframe()`. Adds a shapely point `geometry` attribute to each point in the `gplately.Points` object. pandas.DataFrame that has a column with geometry Any existing point attributes are kept. Returns ------- GeoDataFrame : instance of `geopandas.GeoDataFrame` A pandas.DataFrame with rows equal to the number of points in the `gplately.Points` object, and an additional column containing a shapely `geometry` attribute. Example ------- pt_lons = np.array([140., 150., 160.]) pt_lats = np.array([-30., -40., -50.]) gpts = gplately.Points(model, pt_lons, pt_lats) # Add sample attributes a, b and c to the points in the Points object gpts.add_attributes( a=[10,2,2], b=[2,3,3], c=[30,0,0], ) gpts.get_geopandas_dataframe() ...has the output: a b c geometry 0 10 2 30 POINT (140.00000 -30.00000) 1 2 3 0 POINT (150.00000 -40.00000) 2 2 3 0 POINT (160.00000 -50.00000) """ return self.get_geopandas_dataframe() def get_geopandas_dataframe(self)-
Adds a shapely point
gplately.geometryattribute to each point in thePointsobject. pandas.DataFrame that has a column with geometry Any existing point attributes are kept.Returns
GeoDataFrame:instanceofgeopandas.GeoDataFrame- A pandas.DataFrame with rows equal to the number of points in the
Pointsobject, and an additional column containing a shapelygplately.geometryattribute.
Example
pt_lons = np.array([140., 150., 160.]) pt_lats = np.array([-30., -40., -50.]) gpts = gplately.Points(model, pt_lons, pt_lats) # Add sample attributes a, b and c to the points in the Points object gpts.add_attributes( a=[10,2,2], b=[2,3,3], c=[30,0,0], ) gpts.get_geopandas_dataframe()…has the output:
a b c geometry 0 10 2 30 POINT (140.00000 -30.00000) 1 2 3 0 POINT (150.00000 -40.00000) 2 2 3 0 POINT (160.00000 -50.00000)Expand source code
def get_geopandas_dataframe(self): """Adds a shapely point `geometry` attribute to each point in the `gplately.Points` object. pandas.DataFrame that has a column with geometry Any existing point attributes are kept. Returns ------- GeoDataFrame : instance of `geopandas.GeoDataFrame` A pandas.DataFrame with rows equal to the number of points in the `gplately.Points` object, and an additional column containing a shapely `geometry` attribute. Example ------- pt_lons = np.array([140., 150., 160.]) pt_lats = np.array([-30., -40., -50.]) gpts = gplately.Points(model, pt_lons, pt_lats) # Add sample attributes a, b and c to the points in the Points object gpts.add_attributes( a=[10,2,2], b=[2,3,3], c=[30,0,0], ) gpts.get_geopandas_dataframe() ...has the output: a b c geometry 0 10 2 30 POINT (140.00000 -30.00000) 1 2 3 0 POINT (150.00000 -40.00000) 2 2 3 0 POINT (160.00000 -50.00000) """ import geopandas as gpd from shapely import geometry # create shapely points points = [] for lon, lat in zip(self.lons, self.lats): points.append(geometry.Point(lon, lat)) attributes = self.attributes.copy() attributes["geometry"] = points return gpd.GeoDataFrame(attributes, geometry="geometry") def motion_path(self, time_array, anchor_plate_id=0, return_rate_of_motion=False)-
Create a path of points to mark the trajectory of a plate's motion through geological time.
Parameters
time_array:arr- An array of reconstruction times at which to determine the trajectory
of a point on a plate. For example:
import numpy as np min_time = 30 max_time = 100 time_step = 2.5 time_array = np.arange(min_time, max_time + time_step, time_step) anchor_plate_id:int, default=0- The ID of the anchor plate.
return_rate_of_motion:bool, default=False- Choose whether to return the rate of plate motion through time for each
Returns
rlons:ndarray- An n-dimensional array with columns containing the longitudes of
the seed points at each timestep in
time_array. There are n columns for n seed points. rlats:ndarray- An n-dimensional array with columns containing the latitudes of
the seed points at each timestep in
time_array. There are n columns for n seed points.
Expand source code
def motion_path(self, time_array, anchor_plate_id=0, return_rate_of_motion=False): """Create a path of points to mark the trajectory of a plate's motion through geological time. Parameters ---------- time_array : arr An array of reconstruction times at which to determine the trajectory of a point on a plate. For example: import numpy as np min_time = 30 max_time = 100 time_step = 2.5 time_array = np.arange(min_time, max_time + time_step, time_step) anchor_plate_id : int, default=0 The ID of the anchor plate. return_rate_of_motion : bool, default=False Choose whether to return the rate of plate motion through time for each Returns ------- rlons : ndarray An n-dimensional array with columns containing the longitudes of the seed points at each timestep in `time_array`. There are n columns for n seed points. rlats : ndarray An n-dimensional array with columns containing the latitudes of the seed points at each timestep in `time_array`. There are n columns for n seed points. """ time_array = np.atleast_1d(time_array) # ndarrays to fill with reconstructed points and # rates of motion (if requested) rlons = np.empty((len(time_array), len(self.lons))) rlats = np.empty((len(time_array), len(self.lons))) for i, point_feature in enumerate(self.FeatureCollection): # Create the motion path feature motion_path_feature = pygplates.Feature.create_motion_path( point_feature.get_geometry(), time_array.tolist(), valid_time=(time_array.max(), time_array.min()), # relative_plate=int(self.plate_id[i]), # reconstruction_plate_id=int(anchor_plate_id)) relative_plate=int(self.plate_id[i]), reconstruction_plate_id=int(anchor_plate_id), ) reconstructed_motion_paths = self.plate_reconstruction.reconstruct( motion_path_feature, to_time=0, # from_time=0, reconstruct_type=pygplates.ReconstructType.motion_path, anchor_plate_id=int(anchor_plate_id), ) # Turn motion paths in to lat-lon coordinates for reconstructed_motion_path in reconstructed_motion_paths: trail = reconstructed_motion_path.get_motion_path().to_lat_lon_array() lon, lat = np.flipud(trail[:, 1]), np.flipud(trail[:, 0]) rlons[:, i] = lon rlats[:, i] = lat # Obtain step-plot coordinates for rate of motion if return_rate_of_motion is True: StepTimes = np.empty(((len(time_array) - 1) * 2, len(self.lons))) StepRates = np.empty(((len(time_array) - 1) * 2, len(self.lons))) # Get timestep TimeStep = [] for j in range(len(time_array) - 1): diff = time_array[j + 1] - time_array[j] TimeStep.append(diff) # Iterate over each segment in the reconstructed motion path, get the distance travelled by the moving # plate relative to the fixed plate in each time step Dist = [] for reconstructed_motion_path in reconstructed_motion_paths: for ( segment ) in reconstructed_motion_path.get_motion_path().get_segments(): Dist.append( segment.get_arc_length() * _tools.geocentric_radius( segment.get_start_point().to_lat_lon()[0] ) / 1e3 ) # Note that the motion path coordinates come out starting with the oldest time and working forwards # So, to match our 'times' array, we flip the order Dist = np.flipud(Dist) # Get rate of motion as distance per Myr Rate = np.asarray(Dist) / TimeStep # Manipulate arrays to get a step plot StepRate = np.zeros(len(Rate) * 2) StepRate[::2] = Rate StepRate[1::2] = Rate StepTime = np.zeros(len(Rate) * 2) StepTime[::2] = time_array[:-1] StepTime[1::2] = time_array[1:] # Append the nth point's step time and step rate coordinates to the ndarray StepTimes[:, i] = StepTime StepRates[:, i] = StepRate * 0.1 # cm/yr if return_rate_of_motion is True: return ( np.squeeze(rlons), np.squeeze(rlats), np.squeeze(StepTimes), np.squeeze(StepRates), ) else: return np.squeeze(rlons), np.squeeze(rlats) def plate_velocity(self, time, delta_time=1)-
Calculates the x and y components of tectonic plate velocities at a particular geological time.
This method accesses and uses the
rotation_modelattribute from thePlateReconstructionobject and uses thePointsobject'sself.featuresattribute. Feature points are extracted and assigned plate IDs. These IDs are used to obtain the equivalent stage rotations of identified tectonic plates over a time intervaldelta_time. Each feature point and its stage rotation are used to calculate the point's plate velocity at a particular geological time. Obtained velocities for each domain point are represented in the north-east-down coordinate system, and their x,y Cartesian coordinate components are extracted.Parameters
time:float- The specific geological time (Ma) at which to calculate plate velocities.
delta_time:float, default=1.0- The time increment used for generating partitioning plate stage rotations. 1.0 Ma by default.
Returns
all_velocities.T : 2D numpy list- A transposed 2D numpy list with two rows and a number of columns equal to the number of x,y Cartesian velocity components obtained (and thus the number of feature points extracted from a supplied feature). Each list column stores one point’s x,y, velocity components along its two rows.
Expand source code
def plate_velocity(self, time, delta_time=1): """Calculates the x and y components of tectonic plate velocities at a particular geological time. This method accesses and uses the `rotation_model` attribute from the `PlateReconstruction` object and uses the `Points` object's `self.features` attribute. Feature points are extracted and assigned plate IDs. These IDs are used to obtain the equivalent stage rotations of identified tectonic plates over a time interval `delta_time`. Each feature point and its stage rotation are used to calculate the point's plate velocity at a particular geological time. Obtained velocities for each domain point are represented in the north-east-down coordinate system, and their x,y Cartesian coordinate components are extracted. Parameters ---------- time : float The specific geological time (Ma) at which to calculate plate velocities. delta_time : float, default=1.0 The time increment used for generating partitioning plate stage rotations. 1.0 Ma by default. Returns ------- all_velocities.T : 2D numpy list A transposed 2D numpy list with two rows and a number of columns equal to the number of x,y Cartesian velocity components obtained (and thus the number of feature points extracted from a supplied feature). Each list column stores one point’s x,y, velocity components along its two rows. """ time = float(time) rotation_model = self.plate_reconstruction.rotation_model all_velocities = np.empty((len(self.features), 2)) for i, feature in enumerate(self.features): geometry = feature.get_geometry() partitioning_plate_id = feature.get_reconstruction_plate_id() equivalent_stage_rotation = rotation_model.get_rotation( time, partitioning_plate_id, time + delta_time ) velocity_vectors = pygplates.calculate_velocities( [geometry], equivalent_stage_rotation, delta_time, pygplates.VelocityUnits.cms_per_yr, ) velocities = ( pygplates.LocalCartesian.convert_from_geocentric_to_north_east_down( [geometry], velocity_vectors ) ) all_velocities[i] = velocities[0].get_y(), velocities[0].get_x() return list(all_velocities.T) def reconstruct(self, time, anchor_plate_id=None, return_array=False, **kwargs)-
Reconstructs regular geological features, motion paths or flowlines to a specific geological time and extracts the latitudinal and longitudinal points of these features.
Note: this method accesses and uses the rotation model attribute from the PointReconstruction object, and reconstructs the feature lat-lon point attributes of the Points object.
Parameters
time:float- The specific geological time (Ma) to reconstruct features to.
anchor_plate_id:int, default=None- Reconstruct features with respect to a certain anchor plate. By default, reconstructions are made
with respect to the anchor_plate_ID specified in the
PlateReconstructionobject, which is a default plate ID of 0 unless otherwise specified. return_array:bool, defaultFalse- Return a
numpy.ndarray, rather than aPointsobject. **reconstruct_type:ReconstructType, default=ReconstructType.feature_geometry- The specific reconstruction type to generate based on input feature geometry type. Can be provided as
ReconstructType.feature_geometry to only reconstruct regular feature geometries, or ReconstructType.MotionPath to
only reconstruct motion path features, or ReconstructType.Flowline to only reconstruct flowline features. Generates
:class:
reconstructed feature geometries<ReconstructedFeatureGeometry>’, or :class:reconstructed motion paths’, or :class:`reconstructed flowlines ’ respectively. **group_with_feature:bool, default=False- Used to group reconstructed geometries with their features. This can be useful when a feature has more than one
geometry and hence more than one reconstructed geometry. The output reconstructed_geometries then becomes a
list of tuples where each tuple contains a :class:
feature<Feature>and alistof reconstructed geometries. Note: this keyword argument only applies when reconstructed_geometries is a list because exported files are always grouped with their features. This is applicable to all ReconstructType features. **export_wrap_to_dateline:bool, default=True- Wrap/clip reconstructed geometries to the dateline (currently ignored).
Returns
rlons:listoffloat- A 1D numpy array enclosing all reconstructed point features' longitudes.
rlats:listoffloat- A 1D numpy array enclosing all reconstructed point features' latitudes.
Raises
NotImplementedError- if the starting time for reconstruction
from_timeis not equal to 0.0
Expand source code
def reconstruct(self, time, anchor_plate_id=None, return_array=False, **kwargs): """Reconstructs regular geological features, motion paths or flowlines to a specific geological time and extracts the latitudinal and longitudinal points of these features. Note: this method accesses and uses the rotation model attribute from the PointReconstruction object, and reconstructs the feature lat-lon point attributes of the Points object. Parameters ---------- time : float The specific geological time (Ma) to reconstruct features to. anchor_plate_id : int, default=None Reconstruct features with respect to a certain anchor plate. By default, reconstructions are made with respect to the anchor_plate_ID specified in the `gplately.PlateReconstruction` object, which is a default plate ID of 0 unless otherwise specified. return_array : bool, default False Return a `numpy.ndarray`, rather than a `Points` object. **reconstruct_type : ReconstructType, default=ReconstructType.feature_geometry The specific reconstruction type to generate based on input feature geometry type. Can be provided as ReconstructType.feature_geometry to only reconstruct regular feature geometries, or ReconstructType.MotionPath to only reconstruct motion path features, or ReconstructType.Flowline to only reconstruct flowline features. Generates :class:`reconstructed feature geometries<ReconstructedFeatureGeometry>’, or :class:`reconstructed motion paths<ReconstructedMotionPath>’, or :class:`reconstructed flowlines<ReconstructedFlowline>’ respectively. **group_with_feature : bool, default=False Used to group reconstructed geometries with their features. This can be useful when a feature has more than one geometry and hence more than one reconstructed geometry. The output *reconstructed_geometries* then becomes a list of tuples where each tuple contains a :class:`feature<Feature>` and a ``list`` of reconstructed geometries. Note: this keyword argument only applies when *reconstructed_geometries* is a list because exported files are always grouped with their features. This is applicable to all ReconstructType features. **export_wrap_to_dateline : bool, default=True Wrap/clip reconstructed geometries to the dateline (currently ignored). Returns ------- rlons : list of float A 1D numpy array enclosing all reconstructed point features' longitudes. rlats : list of float A 1D numpy array enclosing all reconstructed point features' latitudes. Raises ------ NotImplementedError if the starting time for reconstruction `from_time` is not equal to 0.0 """ from_time = self.time to_time = time if not anchor_plate_id: anchor_plate_id = self.plate_reconstruction.anchor_plate_id reconstructed_features = self.plate_reconstruction.reconstruct( self.features, to_time, from_time, anchor_plate_id=anchor_plate_id, **kwargs ) rlons, rlats = _tools.extract_feature_lonlat(reconstructed_features) if return_array: return rlons, rlats else: gpts = Points( self.plate_reconstruction, rlons, rlats, time=to_time, plate_id=self.plate_id, ) gpts.add_attributes(**self.attributes.copy()) return gpts def reconstruct_to_birth_age(self, ages, anchor_plate_id=None, **kwargs)-
Reconstructs point features supplied to the
Pointsobject from the supplied initial time (self.time) to a range of times. The number of supplied times must equal the number of point features supplied to the Points object.Attributes
ages:array- Geological times to reconstruct features to. Must have the same length as the
Pointsobject'sself.featuresattribute (which holds all point features represented on a unit length sphere in 3D Cartesian coordinates). anchor_plate_id:int, default=None- Reconstruct features with respect to a certain anchor plate. By default, reconstructions are made
with respect to the anchor_plate_ID specified in the
PlateReconstructionobject, which is a default plate ID of 0 unless otherwise specified. **kwargs- Additional keyword arguments for the
PlateReconstruction.reconstruct()method.
Raises
ValueError- If the number of ages and number of point features supplied to the Points object are not identical.
Returns
rlons,rlats:float- The longitude and latitude coordinate lists of all point features reconstructed to all specified ages.
Examples
To reconstruct n seed points' locations to B Ma (for this example n=2, with (lon,lat) = (78,30) and (56,22) at time=0 Ma, and we reconstruct to B=10 Ma):
# Longitude and latitude of n=2 seed points pt_lon = np.array([78., 56]) pt_lat = np.array([30., 22]) # Call the Points object! gpts = gplately.Points(model, pt_lon, pt_lat) print(gpts.features[0].get_all_geometries()) # Confirms we have features represented as points on a sphere ages = numpy.linspace(10,10, len(pt_lon)) rlons, rlats = gpts.reconstruct_to_birth_age(ages)Expand source code
def reconstruct_to_birth_age(self, ages, anchor_plate_id=None, **kwargs): """Reconstructs point features supplied to the `Points` object from the supplied initial time (`self.time`) to a range of times. The number of supplied times must equal the number of point features supplied to the Points object. Attributes ---------- ages : array Geological times to reconstruct features to. Must have the same length as the `Points `object's `self.features` attribute (which holds all point features represented on a unit length sphere in 3D Cartesian coordinates). anchor_plate_id : int, default=None Reconstruct features with respect to a certain anchor plate. By default, reconstructions are made with respect to the anchor_plate_ID specified in the `gplately.PlateReconstruction` object, which is a default plate ID of 0 unless otherwise specified. **kwargs Additional keyword arguments for the `gplately.PlateReconstruction.reconstruct` method. Raises ------ ValueError If the number of ages and number of point features supplied to the Points object are not identical. Returns ------- rlons, rlats : float The longitude and latitude coordinate lists of all point features reconstructed to all specified ages. Examples -------- To reconstruct n seed points' locations to B Ma (for this example n=2, with (lon,lat) = (78,30) and (56,22) at time=0 Ma, and we reconstruct to B=10 Ma): # Longitude and latitude of n=2 seed points pt_lon = np.array([78., 56]) pt_lat = np.array([30., 22]) # Call the Points object! gpts = gplately.Points(model, pt_lon, pt_lat) print(gpts.features[0].get_all_geometries()) # Confirms we have features represented as points on a sphere ages = numpy.linspace(10,10, len(pt_lon)) rlons, rlats = gpts.reconstruct_to_birth_age(ages) """ from_time = self.time if not anchor_plate_id: anchor_plate_id = self.plate_reconstruction.anchor_plate_id ages = np.array(ages) if len(ages) != len(self.features): raise ValueError("Number of points and ages must be identical") unique_ages = np.unique(ages) rlons = np.zeros(ages.shape) rlats = np.zeros(ages.shape) for age in unique_ages: mask_age = ages == age reconstructed_features = self.plate_reconstruction.reconstruct( self.features, age, from_time, anchor_plate_id=anchor_plate_id, **kwargs ) lons, lats = _tools.extract_feature_lonlat(reconstructed_features) rlons[mask_age] = lons[mask_age] rlats[mask_age] = lats[mask_age] return rlons, rlats def save(self, filename)-
Saves the feature collection used in the Points object under a given filename to the current directory.
The file format is determined from the filename extension.
Parameters
filename:string- Can be provided as a string including the filename and the file format needed.
Returns
Feature collection saved under given filename to current directory.
Expand source code
def save(self, filename): """Saves the feature collection used in the Points object under a given filename to the current directory. The file format is determined from the filename extension. Parameters ---------- filename : string Can be provided as a string including the filename and the file format needed. Returns ------- Feature collection saved under given filename to current directory. """ filename = str(filename) if filename.endswith((".csv", ".txt", ".dat")): df = self._get_dataframe() df.to_csv(filename, index=False) elif filename.endswith((".xls", ".xlsx")): df = self._get_dataframe() df.to_excel(filename, index=False) elif filename.endswith("xml"): df = self._get_dataframe() df.to_xml(filename, index=False) elif filename.endswith(".gpml") or filename.endswith(".gpmlz"): self.FeatureCollection.write(filename) else: raise ValueError( "Cannot save to specified file type. Use csv, gpml, or xls file extension." )
class Raster (data=None, plate_reconstruction=None, extent='global', realign=False, resample=None, time=0.0, origin=None, **kwargs)-
A class for working with raster data.
Raster's functionalities inclue sampling data at points using spline interpolation, resampling rasters with new X and Y-direction spacings and resizing rasters using new X and Y grid pixel resolutions. NaN-type data in rasters can be replaced with the values of their nearest valid neighbours.Parameters
data:strorarray-like- The raster data, either as a filename (
str) or array. plate_reconstruction:PlateReconstruction- Allows for the accessibility of PlateReconstruction object attributes.
Namely, PlateReconstruction object attributes rotation_model,
topology_features and static_polygons can be used in the
Rasterobject if called using “self.plate_reconstruction.X”, where X is the attribute. extent:stror4-tuple, default: 'global'- 4-tuple to specify (min_lon, max_lon, min_lat, max_lat) extents
of the raster. If no extents are supplied, full global extent
[-180,180,-90,90] is assumed (equivalent to
extent='global'). For array data with an upper-left origin, make suremin_latis greater thanmax_lat, or specifyoriginparameter. resample:2-tuple, optional- Optionally resample grid, pass spacing in X and Y direction as a 2-tuple e.g. resample=(spacingX, spacingY).
time:float, default: 0.0- The time step represented by the raster data. Used for raster reconstruction.
origin:{'lower', 'upper'}, optional- When
datais an array, use this parameter to specify the origin (upper left or lower left) of the data (overridingextent). **kwargs- Handle deprecated arguments such as
PlateReconstruction_object,filename, andarray.
Attributes
data:ndarray, shape (ny, nx)- Array containing the underlying raster data. This attribute can be
modified after creation of the
Raster. plate_reconstruction:PlateReconstruction- An object of GPlately's
PlateReconstructionclass, like therotation_model, a set of reconstructabletopology_featuresandstatic_polygonsthat belong to a particular plate model. These attributes can be used in theRasterobject if called using “self.plate_reconstruction.X”, where X is the attribute. This attribute can be modified after creation of theRaster. extent:tupleoffloats- Four-element array to specify [min lon, max lon, min lat, max lat] extents of any sampling points. If no extents are supplied, full global extent [-180,180,-90,90] is assumed.
lons:ndarray, shape (nx,)- The x-coordinates of the raster data. This attribute can be modified
after creation of the
Raster. lats:ndarray, shape (ny,)- The y-coordinates of the raster data. This attribute can be modified
after creation of the
Raster. origin:{'lower', 'upper'}- The origin (lower or upper left) or the data array.
filename:strorNone- The filename used to create the
Rasterobject. If the object was created directly from an array, this attribute isNone.
Methods
interpolate(lons, lats, method='linear', return_indices=False) Sample gridded data at a set of points using spline interpolation.
resample(spacingX, spacingY, overwrite=False) Resamples the grid using X & Y-spaced lat-lon arrays, meshed with linear interpolation.
resize(resX, resY, overwrite=False) Resizes the grid with a specific resolution and samples points using linear interpolation.
fill_NaNs(overwrite=False) Searches for invalid 'data' cells containing NaN-type entries and replaces NaNs with the value of the nearest valid data cell.
reconstruct(time, fill_value=None, partitioning_features=None, threads=1, anchor_plate_id=0, inplace=False) Reconstruct the raster from its initial time (
self.time) to a new time.Constructs all necessary attributes for the raster object.
Note: either a str path to a netCDF file OR an ndarray representing a grid must be specified.
Parameters
data:strorarray-like- The raster data, either as a filename (
str) or array. plate_reconstruction:PlateReconstruction- Allows for the accessibility of PlateReconstruction object attributes. Namely, PlateReconstruction object attributes rotation_model, topology_featues and static_polygons can be used in the points object if called using “self.plate_reconstruction.X”, where X is the attribute.
extent:stror4-tuple, default: 'global'- 4-tuple to specify (min_lon, max_lon, min_lat, max_lat) extents
of the raster. If no extents are supplied, full global extent
[-180,180,-90,90] is assumed (equivalent to
extent='global'). For array data with an upper-left origin, make suremin_latis greater thanmax_lat, or specifyoriginparameter. resample:2-tuple, optional- Optionally resample grid, pass spacing in X and Y direction as a 2-tuple e.g. resample=(spacingX, spacingY).
time:float, default: 0.0- The time step represented by the raster data. Used for raster reconstruction.
origin:{'lower', 'upper'}, optional- When
datais an array, use this parameter to specify the origin (upper left or lower left) of the data (overridingextent). **kwargs- Handle deprecated arguments such as
PlateReconstruction_object,filename, andarray.
Expand source code
class Raster(object): """A class for working with raster data. `Raster`'s functionalities inclue sampling data at points using spline interpolation, resampling rasters with new X and Y-direction spacings and resizing rasters using new X and Y grid pixel resolutions. NaN-type data in rasters can be replaced with the values of their nearest valid neighbours. Parameters ---------- data : str or array-like The raster data, either as a filename (`str`) or array. plate_reconstruction : PlateReconstruction Allows for the accessibility of PlateReconstruction object attributes. Namely, PlateReconstruction object attributes rotation_model, topology_features and static_polygons can be used in the `Raster` object if called using “self.plate_reconstruction.X”, where X is the attribute. extent : str or 4-tuple, default: 'global' 4-tuple to specify (min_lon, max_lon, min_lat, max_lat) extents of the raster. If no extents are supplied, full global extent [-180,180,-90,90] is assumed (equivalent to `extent='global'`). For array data with an upper-left origin, make sure `min_lat` is greater than `max_lat`, or specify `origin` parameter. resample : 2-tuple, optional Optionally resample grid, pass spacing in X and Y direction as a 2-tuple e.g. resample=(spacingX, spacingY). time : float, default: 0.0 The time step represented by the raster data. Used for raster reconstruction. origin : {'lower', 'upper'}, optional When `data` is an array, use this parameter to specify the origin (upper left or lower left) of the data (overriding `extent`). **kwargs Handle deprecated arguments such as `PlateReconstruction_object`, `filename`, and `array`. Attributes ---------- data : ndarray, shape (ny, nx) Array containing the underlying raster data. This attribute can be modified after creation of the `Raster`. plate_reconstruction : PlateReconstruction An object of GPlately's `PlateReconstruction` class, like the `rotation_model`, a set of reconstructable `topology_features` and `static_polygons` that belong to a particular plate model. These attributes can be used in the `Raster` object if called using “self.plate_reconstruction.X”, where X is the attribute. This attribute can be modified after creation of the `Raster`. extent : tuple of floats Four-element array to specify [min lon, max lon, min lat, max lat] extents of any sampling points. If no extents are supplied, full global extent [-180,180,-90,90] is assumed. lons : ndarray, shape (nx,) The x-coordinates of the raster data. This attribute can be modified after creation of the `Raster`. lats : ndarray, shape (ny,) The y-coordinates of the raster data. This attribute can be modified after creation of the `Raster`. origin : {'lower', 'upper'} The origin (lower or upper left) or the data array. filename : str or None The filename used to create the `Raster` object. If the object was created directly from an array, this attribute is `None`. Methods ------- interpolate(lons, lats, method='linear', return_indices=False) Sample gridded data at a set of points using spline interpolation. resample(spacingX, spacingY, overwrite=False) Resamples the grid using X & Y-spaced lat-lon arrays, meshed with linear interpolation. resize(resX, resY, overwrite=False) Resizes the grid with a specific resolution and samples points using linear interpolation. fill_NaNs(overwrite=False) Searches for invalid 'data' cells containing NaN-type entries and replaces NaNs with the value of the nearest valid data cell. reconstruct(time, fill_value=None, partitioning_features=None, threads=1, anchor_plate_id=0, inplace=False) Reconstruct the raster from its initial time (`self.time`) to a new time. """ def __init__( self, data=None, plate_reconstruction=None, extent="global", realign=False, resample=None, time=0.0, origin=None, **kwargs ): """Constructs all necessary attributes for the raster object. Note: either a str path to a netCDF file OR an ndarray representing a grid must be specified. Parameters ---------- data : str or array-like The raster data, either as a filename (`str`) or array. plate_reconstruction : PlateReconstruction Allows for the accessibility of PlateReconstruction object attributes. Namely, PlateReconstruction object attributes rotation_model, topology_featues and static_polygons can be used in the points object if called using “self.plate_reconstruction.X”, where X is the attribute. extent : str or 4-tuple, default: 'global' 4-tuple to specify (min_lon, max_lon, min_lat, max_lat) extents of the raster. If no extents are supplied, full global extent [-180,180,-90,90] is assumed (equivalent to `extent='global'`). For array data with an upper-left origin, make sure `min_lat` is greater than `max_lat`, or specify `origin` parameter. resample : 2-tuple, optional Optionally resample grid, pass spacing in X and Y direction as a 2-tuple e.g. resample=(spacingX, spacingY). time : float, default: 0.0 The time step represented by the raster data. Used for raster reconstruction. origin : {'lower', 'upper'}, optional When `data` is an array, use this parameter to specify the origin (upper left or lower left) of the data (overriding `extent`). **kwargs Handle deprecated arguments such as `PlateReconstruction_object`, `filename`, and `array`. """ if isinstance(data, self.__class__): self._data = data._data.copy() self.plate_reconstruction = data.plate_reconstruction self._lons = data._lons self._lats = data._lats self._time = data._time return if "PlateReconstruction_object" in kwargs.keys(): warnings.warn( "`PlateReconstruction_object` keyword argument has been " + "deprecated, use `plate_reconstruction` instead", DeprecationWarning, ) if plate_reconstruction is None: plate_reconstruction = kwargs.pop("PlateReconstruction_object") if "filename" in kwargs.keys() and "array" in kwargs.keys(): raise TypeError( "Both `filename` and `array` were provided; use " + "one or the other, or use the `data` argument" ) if "filename" in kwargs.keys(): warnings.warn( "`filename` keyword argument has been deprecated, " + "use `data` instead", DeprecationWarning, ) if data is None: data = kwargs.pop("filename") if "array" in kwargs.keys(): warnings.warn( "`array` keyword argument has been deprecated, " + "use `data` instead", DeprecationWarning, ) if data is None: data = kwargs.pop("array") for key in kwargs.keys(): raise TypeError( "Raster.__init__() got an unexpected keyword argument " + "'{}'".format(key) ) self.plate_reconstruction = plate_reconstruction if time < 0.0: raise ValueError("Invalid time: {}".format(time)) time = float(time) self._time = time if data is None: raise TypeError( "`data` argument (or `filename` or `array`) is required" ) if isinstance(data, str): # Filename self._filename = data self._data, lons, lats = read_netcdf_grid( data, return_grids=True, realign=realign, resample=resample, ) self._lons = lons self._lats = lats else: # numpy array self._filename = None extent = _parse_extent_origin(extent, origin) data = _check_grid(data) self._data = np.array(data) self._lons = np.linspace(extent[0], extent[1], self.data.shape[1]) self._lats = np.linspace(extent[2], extent[3], self.data.shape[0]) if realign: # realign to -180,180 and flip grid self._data, self._lons, self._lats = realign_grid(self._data, self._lons, self._lats) if (not isinstance(data, str)) and (resample is not None): self.resample(*resample, inplace=True) @property def time(self): """The time step represented by the raster data.""" return self._time @property def data(self): """The object's raster data. Can be modified. """ return self._data @data.setter def data(self, z): z = np.array(z) if z.shape != np.shape(self.data): raise ValueError( "Shape mismatch: old dimensions are {}, new are {}".format( np.shape(self.data), z.shape, ) ) self._data = z @property def lons(self): """The x-coordinates of the raster data. Can be modified. """ return self._lons @lons.setter def lons(self, x): x = np.array(x).ravel() if x.size != np.shape(self.data)[1]: raise ValueError( "Shape mismatch: data x-dimension is {}, new value is {}".format( np.shape(self.data)[1], x.size, ) ) self._lons = x @property def lats(self): """The y-coordinates of the raster data. Can be modified. """ return self._lats @lats.setter def lats(self, y): y = np.array(y).ravel() if y.size != np.shape(self.data)[0]: raise ValueError( "Shape mismatch: data y-dimension is {}, new value is {}".format( np.shape(self.data)[0], y.size, ) ) self._lats = y @property def extent(self): """The spatial extent (x0, x1, y0, y1) of the data. If y0 < y1, the origin is the lower-left corner; else the upper-left. """ return ( float(self.lons[0]), float(self.lons[-1]), float(self.lats[0]), float(self.lats[-1]), ) @property def origin(self): """The origin of the data array, used for e.g. plotting.""" if self.lats[0] < self.lats[-1]: return "lower" else: return "upper" @property def shape(self): """The shape of the data array.""" return np.shape(self.data) @property def size(self): """The size of the data array.""" return np.size(self.data) @property def dtype(self): """The data type of the array.""" return self.data.dtype @property def ndim(self): """The number of dimensions in the array.""" return np.ndim(self.data) @property def filename(self): """The filename of the raster file used to create the object. If a NumPy array was used instead, this attribute is `None`. """ return self._filename @property def plate_reconstruction(self): """The `PlateReconstruction` object to be used for raster reconstruction. """ return self._plate_reconstruction @plate_reconstruction.setter def plate_reconstruction(self, reconstruction): if reconstruction is None: # Remove `plate_reconstruction` attribute pass elif not isinstance(reconstruction, _PlateReconstruction): # Convert to a `PlateReconstruction` if possible try: reconstruction = _PlateReconstruction(*reconstruction) except Exception: reconstruction = _PlateReconstruction(reconstruction) self._plate_reconstruction = reconstruction def copy(self): """ Returns a copy of the Raster Returns ------- Raster A copy of the current Raster object """ return Raster(self.data.copy(), self.plate_reconstruction, self.extent, self.time) def interpolate( self, lons, lats, method="linear", return_indices=False, ): """Interpolate a set of point data onto the gridded data provided to the `Raster` object. Parameters ---------- lons, lats : array_like The longitudes and latitudes of the points to interpolate onto the gridded data. Must be broadcastable to a common shape. method : str or int; default: 'linear' The order of spline interpolation. Must be an integer in the range 0-5. 'nearest', 'linear', and 'cubic' are aliases for 0, 1, and 3, respectively. return_indices : bool, default=False Whether to return the row and column indices of the nearest grid points. Returns ------- numpy.ndarray The values interpolated at the input points. indices : 2-tuple of numpy.ndarray The i- and j-indices of the nearest grid points to the input points, only present if `return_indices=True`. Raises ------ ValueError If an invalid `method` is provided. RuntimeWarning If `lats` contains any invalid values outside of the interval [-90, 90]. Invalid values will be clipped to this interval. Notes ----- If `return_indices` is set to `True`, the nearest array indices are returned as a tuple of arrays, in (i, j) or (lat, lon) format. An example output: # The first array holds the rows of the raster where point data spatially falls near. # The second array holds the columns of the raster where point data spatially falls near. sampled_indices = (array([1019, 1019, 1019, ..., 1086, 1086, 1087]), array([2237, 2237, 2237, ..., 983, 983, 983])) """ return sample_grid( lon=lons, lat=lats, grid=self, method=method, return_indices=return_indices, ) def resample(self, spacingX, spacingY, method="linear", inplace=False): """Resample the `grid` passed to the `Raster` object with a new `spacingX` and `spacingY` using linear interpolation. Notes ----- Ultimately, `resample` changes the lat-lon resolution of the gridded data. The larger the x and y spacings given are, the larger the pixellation of raster data. `resample` creates new latitude and longitude arrays with specified spacings in the X and Y directions (`spacingX` and `spacingY`). These arrays are linearly interpolated into a new raster. If `inplace` is set to `True`, the respaced latitude array, longitude array and raster will inplace the ones currently attributed to the `Raster` object. Parameters ---------- spacingX, spacingY : ndarray Specify the spacing in the X and Y directions with which to resample. The larger `spacingX` and `spacingY` are, the larger the raster pixels become (less resolved). Note: to keep the size of the raster consistent, set `spacingX = spacingY`; otherwise, if for example `spacingX > spacingY`, the raster will appear stretched longitudinally. method : str or int; default: 'linear' The order of spline interpolation. Must be an integer in the range 0-5. 'nearest', 'linear', and 'cubic' are aliases for 0, 1, and 3, respectively. inplace : bool, default=False Choose to overwrite the data (the `self.data` attribute), latitude array (`self.lats`) and longitude array (`self.lons`) currently attributed to the `Raster` object. Returns ------- Raster The resampled grid. If `inplace` is set to `True`, this raster overwrites the one attributed to `data`. """ spacingX = np.abs(spacingX) spacingY = np.abs(spacingY) if self.origin == "upper": spacingY *= -1.0 lons = np.arange(self.extent[0], self.extent[1]+spacingX, spacingX) lats = np.arange(self.extent[2], self.extent[3]+spacingY, spacingY) lonq, latq = np.meshgrid(lons, lats) data = self.interpolate(lonq, latq, method=method) if inplace: self._data = data self._lons = lons self._lats = lats else: return Raster(data, self.plate_reconstruction, self.extent, self.time) def resize(self, resX, resY, inplace=False, method="linear", return_array=False): """Resize the grid passed to the `Raster` object with a new x and y resolution (`resX` and `resY`) using linear interpolation. Notes ----- Ultimately, `resize` "stretches" a raster in the x and y directions. The larger the resolutions in x and y, the more stretched the raster appears in x and y. It creates new latitude and longitude arrays with specific resolutions in the X and Y directions (`resX` and `resY`). These arrays are linearly interpolated into a new raster. If `inplace` is set to `True`, the resized latitude, longitude arrays and raster will inplace the ones currently attributed to the `Raster` object. Parameters ---------- resX, resY : ndarray Specify the resolutions with which to resize the raster. The larger `resX` is, the more longitudinally-stretched the raster becomes. The larger `resY` is, the more latitudinally-stretched the raster becomes. method : str or int; default: 'linear' The order of spline interpolation. Must be an integer in the range 0-5. 'nearest', 'linear', and 'cubic' are aliases for 0, 1, and 3, respectively. inplace : bool, default=False Choose to overwrite the data (the `self.data` attribute), latitude array (`self.lats`) and longitude array (`self.lons`) currently attributed to the `Raster` object. return_array : bool, default False Return a `numpy.ndarray`, rather than a `Raster`. Returns ------- Raster The resized grid. If `inplace` is set to `True`, this raster overwrites the one attributed to `data`. """ # construct grid lons = np.linspace(self.extent[0], self.extent[1], resX) lats = np.linspace(self.extent[2], self.extent[3], resY) lonq, latq = np.meshgrid(lons, lats) data = self.interpolate(lonq, latq, method=method) if inplace: self._data = data self._lons = lons self._lats = lats if return_array: return data else: return Raster(data, self.plate_reconstruction, self.extent, self.time) def fill_NaNs(self, inplace=False, return_array=False): """Search raster for invalid ‘data’ cells containing NaN-type entries replaces them with the value of their nearest valid data cells. Parameters --------- inplace : bool, default=False Choose whether to overwrite the grid currently held in the `data` attribute with the filled grid. return_array : bool, default False Return a `numpy.ndarray`, rather than a `Raster`. Returns -------- Raster The resized grid. If `inplace` is set to `True`, this raster overwrites the one attributed to `data`. """ data = fill_raster(self.data) if inplace: self._data = data if return_array: return data else: return Raster(data, self.plate_reconstruction, self.extent, self.time) def save_to_netcdf4(self, filename): """ Saves the grid attributed to the `Raster` object to the given `filename` (including the ".nc" extension) in netCDF4 format.""" write_netcdf_grid(str(filename), self.data, self.extent) def reconstruct( self, time, fill_value=None, partitioning_features=None, threads=1, anchor_plate_id=0, inplace=False, return_array=False, ): """Reconstruct raster data to a given time. Parameters ---------- time : float Time to which the data will be reconstructed. fill_value : float, int, str, or tuple, optional The value to be used for regions outside of the static polygons at `time`. By default (`fill_value=None`), this value will be determined based on the input. partitioning_features : sequence of Feature or str, optional The features used to partition the raster grid and assign plate IDs. By default, `self.plate_reconstruction.static_polygons` will be used, but alternatively any valid argument to `pygplates.FeaturesFunctionArgument` can be specified here. threads : int, default 1 Number of threads to use for certain computationally heavy routines. anchor_plate_id : int, default 0 ID of the anchored plate. inplace : bool, default False Perform the reconstruction in-place (replace the raster's data with the reconstructed data). return_array : bool, default False Return a `numpy.ndarray`, rather than a `Raster`. Returns ------- Raster or np.ndarray The reconstructed grid. Areas for which no plate ID could be determined will be filled with `fill_value`. Raises ------ TypeError If this `Raster` has no `plate_reconstruction` set. Notes ----- For two-dimensional grids, `fill_value` should be a single number. The default value will be `np.nan` for float or complex types, the minimum value for integer types, and the maximum value for unsigned types. For RGB image grids, `fill_value` should be a 3-tuple RGB colour code or a matplotlib colour string. The default value will be black (0.0, 0.0, 0.0) or (0, 0, 0). For RGBA image grids, `fill_value` should be a 4-tuple RGBA colour code or a matplotlib colour string. The default fill value will be transparent black (0.0, 0.0, 0.0, 0.0) or (0, 0, 0, 0). """ if time < 0.0: raise ValueError("Invalid time: {}".format(time)) time = float(time) if self.plate_reconstruction is None: raise TypeError( "Cannot perform reconstruction - " + "`plate_reconstruction` has not been set" ) if partitioning_features is None: partitioning_features = self.plate_reconstruction.static_polygons result = reconstruct_grid( grid=self.data, partitioning_features=partitioning_features, rotation_model=self.plate_reconstruction.rotation_model, from_time=self.time, to_time=time, extent=self.extent, origin=self.origin, fill_value=fill_value, threads=threads, anchor_plate_id=anchor_plate_id, ) if inplace: self.data = result self._time = time if return_array: return result return self if not return_array: result = type(self)( data=result, plate_reconstruction=self.plate_reconstruction, extent=self.extent, time=time, origin=self.origin, ) return result def imshow(self, ax=None, projection=None, **kwargs): """Display raster data. A pre-existing matplotlib `Axes` instance is used if available, else a new one is created. The `origin` and `extent` of the image are determined automatically and should not be specified. Parameters ---------- ax : matplotlib.axes.Axes, optional If specified, the image will be drawn within these axes. projection : cartopy.crs.Projection, optional The map projection to be used. If both `ax` and `projection` are specified, this will be checked against the `projection` attribute of `ax`, if it exists. **kwargs : dict, optional Any further keyword arguments are passed to `matplotlib.pyplot.imshow` or `matplotlib.axes.Axes.imshow`, where appropriate. Returns ------- matplotlib.image.AxesImage Raises ------ ValueError If `ax` and `projection` are both specified, but do not match (i.e. `ax.projection != projection`). """ for kw in ("origin", "extent"): if kw in kwargs.keys(): raise TypeError( "imshow got an unexpected keyword argument: {}".format(kw) ) if ax is None: existing_figure = len(plt.get_fignums()) > 0 current_axes = plt.gca() if projection is None: ax = current_axes elif ( isinstance(current_axes, _GeoAxes) and current_axes.projection == projection ): ax = current_axes else: if not existing_figure: current_axes.remove() ax = plt.axes(projection=projection) elif projection is not None: # projection and ax both specified if isinstance(ax, _GeoAxes) and ax.projection == projection: pass # projections match else: raise ValueError( "Both `projection` and `ax` were specified, but" + " `projection` does not match `ax.projection`" ) if isinstance(ax, _GeoAxes) and "transform" not in kwargs.keys(): kwargs["transform"] = _PlateCarree() extent = self.extent if self.origin == "upper": extent = ( extent[0], extent[1], extent[3], extent[2], ) im = ax.imshow(self.data, origin=self.origin, extent=extent, **kwargs) return im plot = imshow def rotate_reference_frames( self, grid_spacing_degrees, reconstruction_time, from_rotation_features_or_model, # filename(s), or pyGPlates feature(s)/collection(s) or a RotationModel to_rotation_features_or_model, # filename(s), or pyGPlates feature(s)/collection(s) or a RotationModel from_rotation_reference_plate=0, to_rotation_reference_plate=0, non_reference_plate=701, output_name=None ): import time as timer """Rotate a grid defined in one plate model reference frame within a gplately.Raster object to another plate reconstruction model reference frame. Parameters ---------- grid_spacing_degrees : float The spacing (in degrees) for the output rotated grid. reconstruction_time : float The time at which to rotate the input grid. from_rotation_features_or_model : str, list of str, or instance of pygplates.RotationModel A filename, or a list of filenames, or a pyGPlates RotationModel object that defines the rotation model that the input grid is currently associated with. to_rotation_features_or_model : str, list of str, or instance of pygplates.RotationModel A filename, or a list of filenames, or a pyGPlates RotationModel object that defines the rotation model that the input grid shall be rotated with. from_rotation_reference_plate : int, default = 0 The current reference plate for the plate model the grid is defined in. Defaults to the anchor plate 0. to_rotation_reference_plate : int, default = 0 The desired reference plate for the plate model the grid is being rotated to. Defaults to the anchor plate 0. non_reference_plate : int, default = 701 An arbitrary placeholder reference frame with which to define the "from" and "to" reference frames. output_name : str, default None If passed, the rotated grid is saved as a netCDF grid to this filename. Returns ------- gplately.Raster() An instance of the gplately.Raster object containing the rotated grid. """ input_positions = [] # Create the pygplates.FiniteRotation that rotates # between the two reference frames. from_rotation_model = pygplates.RotationModel( from_rotation_features_or_model ) to_rotation_model = pygplates.RotationModel( to_rotation_features_or_model ) from_rotation = from_rotation_model.get_rotation( reconstruction_time, non_reference_plate, anchor_plate_id=from_rotation_reference_plate ) to_rotation = to_rotation_model.get_rotation( reconstruction_time, non_reference_plate, anchor_plate_id=to_rotation_reference_plate ) reference_frame_conversion_rotation = to_rotation * from_rotation.get_inverse() # Resize the input grid to the specified output resolution before rotating resX = _deg2pixels( grid_spacing_degrees, self.extent[0], self.extent[1] ) resY = _deg2pixels( grid_spacing_degrees, self.extent[2], self.extent[3] ) resized_input_grid = self.resize( resX, resY, inplace=False ) # Get the flattened lons, lats llons, llats = np.meshgrid( resized_input_grid.lons, resized_input_grid.lats ) llons = llons.flatten() llats = llats.flatten() input_coords = [(llons[i], llats[i]) for i in range(0, len(llons))] # Convert lon-lat points of Raster grid to pyGPlates points input_points = pygplates.MultiPointOnSphere( (lat, lon) for lon, lat in input_coords ) # Get grid values of the resized Raster object values = np.array(resized_input_grid.data).flatten() # Rotate grid nodes to the other reference frame output_points = reference_frame_conversion_rotation * input_points # Assemble rotated points with grid values. out_lon = [] out_lat = [] zdata = [] for index, point in enumerate(output_points): lat, lon = point.to_lat_lon() out_lon.append(lon) out_lat.append(lat) zdata.append(values[index]) # Create a regular grid on which to interpolate lats, lons and zdata # Use the extent of the original Raster object extent_globe = self.extent resX = int(np.floor((extent_globe[1] - extent_globe[0]) / grid_spacing_degrees)) + 1 resY = int(np.floor((extent_globe[3] - extent_globe[2]) / grid_spacing_degrees)) + 1 grid_lon = np.linspace( extent_globe[0], extent_globe[1], resX ) grid_lat = np.linspace( extent_globe[2], extent_globe[3], resY ) X, Y = np.meshgrid( grid_lon, grid_lat ) # Interpolate lons, lats and zvals over a regular grid using nearest # neighbour interpolation Z = griddata( (out_lon, out_lat), zdata, (X, Y), method='nearest' ) # Write output grid to netCDF if requested. if output_name: write_netcdf_grid( output_name, Z, extent=extent_globe, ) return Raster(data=Z) def __array__(self): return np.array(self.data) def __add__(self, other): if isinstance(other, Raster): # Return array, since we don't know which Raster # to take properties from return self.data + other.data # Return Raster with new data new_raster = self.copy() new_data = self.data + other new_raster.data = new_data return new_raster def __radd__(self, other): return self + other def __sub__(self, other): if isinstance(other, Raster): # Return array, since we don't know which Raster # to take properties from return self.data - other.data # Return Raster with new data new_raster = self.copy() new_data = self.data - other new_raster.data = new_data return new_raster def __rsub__(self, other): if isinstance(other, Raster): # Return array, since we don't know which Raster # to take properties from return other.data - self.data # Return Raster with new data new_raster = self.copy() new_data = other - self.data new_raster.data = new_data return new_raster def __mul__(self, other): if isinstance(other, Raster): # Return array, since we don't know which Raster # to take properties from return self.data * other.data # Return Raster with new data new_raster = self.copy() new_data = self.data * other new_raster.data = new_data return new_raster def __rmul__(self, other): return self * other def __truediv__(self, other): if isinstance(other, Raster): # Return array, since we don't know which Raster # to take properties from return self.data / other.data # Return Raster with new data new_raster = self.copy() new_data = self.data / other new_raster.data = new_data return new_raster def __rtruediv__(self, other): if isinstance(other, Raster): # Return array, since we don't know which Raster # to take properties from return other.data / self.data # Return Raster with new data new_raster = self.copy() new_data = other / self.data new_raster.data = new_data return new_raster def __floordiv__(self, other): if isinstance(other, Raster): # Return array, since we don't know which Raster # to take properties from return self.data // other.data # Return Raster with new data new_raster = self.copy() new_data = self.data // other new_raster.data = new_data return new_raster def __rfloordiv__(self, other): if isinstance(other, Raster): # Return array, since we don't know which Raster # to take properties from return other.data // self.data # Return Raster with new data new_raster = self.copy() new_data = other // self.data new_raster.data = new_data return new_raster def __mod__(self, other): if isinstance(other, Raster): # Return array, since we don't know which Raster # to take properties from return self.data % other.data # Return Raster with new data new_raster = self.copy() new_data = self.data % other new_raster.data = new_data return new_raster def __rmod__(self, other): if isinstance(other, Raster): # Return array, since we don't know which Raster # to take properties from return other.data % self.data # Return Raster with new data new_raster = self.copy() new_data = other % self.data new_raster.data = new_data return new_raster def __pow__(self, other): if isinstance(other, Raster): # Return array, since we don't know which Raster # to take properties from return self.data ** other.data # Return Raster with new data new_raster = self.copy() new_data = self.data ** other new_raster.data = new_data return new_raster def __rpow__(self, other): if isinstance(other, Raster): # Return array, since we don't know which Raster # to take properties from return other.data ** self.data # Return Raster with new data new_raster = self.copy() new_data = other ** self.data new_raster.data = new_data return new_rasterInstance variables
var data-
The object's raster data.
Can be modified.
Expand source code
@property def data(self): """The object's raster data. Can be modified. """ return self._data var dtype-
The data type of the array.
Expand source code
@property def dtype(self): """The data type of the array.""" return self.data.dtype var extent-
The spatial extent (x0, x1, y0, y1) of the data.
If y0 < y1, the origin is the lower-left corner; else the upper-left.
Expand source code
@property def extent(self): """The spatial extent (x0, x1, y0, y1) of the data. If y0 < y1, the origin is the lower-left corner; else the upper-left. """ return ( float(self.lons[0]), float(self.lons[-1]), float(self.lats[0]), float(self.lats[-1]), ) var filename-
The filename of the raster file used to create the object.
If a NumPy array was used instead, this attribute is
None.Expand source code
@property def filename(self): """The filename of the raster file used to create the object. If a NumPy array was used instead, this attribute is `None`. """ return self._filename var lats-
The y-coordinates of the raster data.
Can be modified.
Expand source code
@property def lats(self): """The y-coordinates of the raster data. Can be modified. """ return self._lats var lons-
The x-coordinates of the raster data.
Can be modified.
Expand source code
@property def lons(self): """The x-coordinates of the raster data. Can be modified. """ return self._lons var ndim-
The number of dimensions in the array.
Expand source code
@property def ndim(self): """The number of dimensions in the array.""" return np.ndim(self.data) var origin-
The origin of the data array, used for e.g. plotting.
Expand source code
@property def origin(self): """The origin of the data array, used for e.g. plotting.""" if self.lats[0] < self.lats[-1]: return "lower" else: return "upper" var plate_reconstruction-
The
PlateReconstructionobject to be used for raster reconstruction.Expand source code
@property def plate_reconstruction(self): """The `PlateReconstruction` object to be used for raster reconstruction. """ return self._plate_reconstruction var shape-
The shape of the data array.
Expand source code
@property def shape(self): """The shape of the data array.""" return np.shape(self.data) var size-
The size of the data array.
Expand source code
@property def size(self): """The size of the data array.""" return np.size(self.data) var time-
The time step represented by the raster data.
Expand source code
@property def time(self): """The time step represented by the raster data.""" return self._time
Methods
def copy(self)-
Expand source code
def copy(self): """ Returns a copy of the Raster Returns ------- Raster A copy of the current Raster object """ return Raster(self.data.copy(), self.plate_reconstruction, self.extent, self.time) def fill_NaNs(self, inplace=False, return_array=False)-
Search raster for invalid ‘data’ cells containing NaN-type entries replaces them with the value of their nearest valid data cells.
Parameters
inplace:bool, default=False- Choose whether to overwrite the grid currently held in the
dataattribute with the filled grid. return_array:bool, defaultFalse- Return a
numpy.ndarray, rather than aRaster.
Returns
Raster- The resized grid. If
inplaceis set toTrue, this raster overwrites the one attributed todata.
Expand source code
def fill_NaNs(self, inplace=False, return_array=False): """Search raster for invalid ‘data’ cells containing NaN-type entries replaces them with the value of their nearest valid data cells. Parameters --------- inplace : bool, default=False Choose whether to overwrite the grid currently held in the `data` attribute with the filled grid. return_array : bool, default False Return a `numpy.ndarray`, rather than a `Raster`. Returns -------- Raster The resized grid. If `inplace` is set to `True`, this raster overwrites the one attributed to `data`. """ data = fill_raster(self.data) if inplace: self._data = data if return_array: return data else: return Raster(data, self.plate_reconstruction, self.extent, self.time) def imshow(self, ax=None, projection=None, **kwargs)-
Display raster data.
A pre-existing matplotlib
Axesinstance is used if available, else a new one is created. Theoriginandextentof the image are determined automatically and should not be specified.Parameters
ax:matplotlib.axes.Axes, optional- If specified, the image will be drawn within these axes.
projection:cartopy.crs.Projection, optional- The map projection to be used. If both
axandprojectionare specified, this will be checked against theprojectionattribute ofax, if it exists. **kwargs:dict, optional- Any further keyword arguments are passed to
matplotlib.pyplot.imshowormatplotlib.axes.Axes.imshow, where appropriate.
Returns
matplotlib.image.AxesImage
Raises
ValueError- If
axandprojectionare both specified, but do not match (i.e.ax.projection != projection).
Expand source code
def imshow(self, ax=None, projection=None, **kwargs): """Display raster data. A pre-existing matplotlib `Axes` instance is used if available, else a new one is created. The `origin` and `extent` of the image are determined automatically and should not be specified. Parameters ---------- ax : matplotlib.axes.Axes, optional If specified, the image will be drawn within these axes. projection : cartopy.crs.Projection, optional The map projection to be used. If both `ax` and `projection` are specified, this will be checked against the `projection` attribute of `ax`, if it exists. **kwargs : dict, optional Any further keyword arguments are passed to `matplotlib.pyplot.imshow` or `matplotlib.axes.Axes.imshow`, where appropriate. Returns ------- matplotlib.image.AxesImage Raises ------ ValueError If `ax` and `projection` are both specified, but do not match (i.e. `ax.projection != projection`). """ for kw in ("origin", "extent"): if kw in kwargs.keys(): raise TypeError( "imshow got an unexpected keyword argument: {}".format(kw) ) if ax is None: existing_figure = len(plt.get_fignums()) > 0 current_axes = plt.gca() if projection is None: ax = current_axes elif ( isinstance(current_axes, _GeoAxes) and current_axes.projection == projection ): ax = current_axes else: if not existing_figure: current_axes.remove() ax = plt.axes(projection=projection) elif projection is not None: # projection and ax both specified if isinstance(ax, _GeoAxes) and ax.projection == projection: pass # projections match else: raise ValueError( "Both `projection` and `ax` were specified, but" + " `projection` does not match `ax.projection`" ) if isinstance(ax, _GeoAxes) and "transform" not in kwargs.keys(): kwargs["transform"] = _PlateCarree() extent = self.extent if self.origin == "upper": extent = ( extent[0], extent[1], extent[3], extent[2], ) im = ax.imshow(self.data, origin=self.origin, extent=extent, **kwargs) return im def interpolate(self, lons, lats, method='linear', return_indices=False)-
Interpolate a set of point data onto the gridded data provided to the
Rasterobject.Parameters
lons,lats:array_like- The longitudes and latitudes of the points to interpolate onto the gridded data. Must be broadcastable to a common shape.
method:strorint; default: 'linear'- The order of spline interpolation. Must be an integer in the range 0-5. 'nearest', 'linear', and 'cubic' are aliases for 0, 1, and 3, respectively.
return_indices:bool, default=False- Whether to return the row and column indices of the nearest grid points.
Returns
numpy.ndarray- The values interpolated at the input points.
indices:2-tupleofnumpy.ndarray- The i- and j-indices of the nearest grid points to the input
points, only present if
return_indices=True.
Raises
ValueError- If an invalid
methodis provided. RuntimeWarning- If
latscontains any invalid values outside of the interval [-90, 90]. Invalid values will be clipped to this interval.
Notes
If
return_indicesis set toTrue, the nearest array indices are returned as a tuple of arrays, in (i, j) or (lat, lon) format.An example output:
# The first array holds the rows of the raster where point data spatially falls near. # The second array holds the columns of the raster where point data spatially falls near. sampled_indices = (array([1019, 1019, 1019, ..., 1086, 1086, 1087]), array([2237, 2237, 2237, ..., 983, 983, 983]))Expand source code
def interpolate( self, lons, lats, method="linear", return_indices=False, ): """Interpolate a set of point data onto the gridded data provided to the `Raster` object. Parameters ---------- lons, lats : array_like The longitudes and latitudes of the points to interpolate onto the gridded data. Must be broadcastable to a common shape. method : str or int; default: 'linear' The order of spline interpolation. Must be an integer in the range 0-5. 'nearest', 'linear', and 'cubic' are aliases for 0, 1, and 3, respectively. return_indices : bool, default=False Whether to return the row and column indices of the nearest grid points. Returns ------- numpy.ndarray The values interpolated at the input points. indices : 2-tuple of numpy.ndarray The i- and j-indices of the nearest grid points to the input points, only present if `return_indices=True`. Raises ------ ValueError If an invalid `method` is provided. RuntimeWarning If `lats` contains any invalid values outside of the interval [-90, 90]. Invalid values will be clipped to this interval. Notes ----- If `return_indices` is set to `True`, the nearest array indices are returned as a tuple of arrays, in (i, j) or (lat, lon) format. An example output: # The first array holds the rows of the raster where point data spatially falls near. # The second array holds the columns of the raster where point data spatially falls near. sampled_indices = (array([1019, 1019, 1019, ..., 1086, 1086, 1087]), array([2237, 2237, 2237, ..., 983, 983, 983])) """ return sample_grid( lon=lons, lat=lats, grid=self, method=method, return_indices=return_indices, ) def plot(self, ax=None, projection=None, **kwargs)-
Display raster data.
A pre-existing matplotlib
Axesinstance is used if available, else a new one is created. Theoriginandextentof the image are determined automatically and should not be specified.Parameters
ax:matplotlib.axes.Axes, optional- If specified, the image will be drawn within these axes.
projection:cartopy.crs.Projection, optional- The map projection to be used. If both
axandprojectionare specified, this will be checked against theprojectionattribute ofax, if it exists. **kwargs:dict, optional- Any further keyword arguments are passed to
matplotlib.pyplot.imshowormatplotlib.axes.Axes.imshow, where appropriate.
Returns
matplotlib.image.AxesImage
Raises
ValueError- If
axandprojectionare both specified, but do not match (i.e.ax.projection != projection).
Expand source code
def imshow(self, ax=None, projection=None, **kwargs): """Display raster data. A pre-existing matplotlib `Axes` instance is used if available, else a new one is created. The `origin` and `extent` of the image are determined automatically and should not be specified. Parameters ---------- ax : matplotlib.axes.Axes, optional If specified, the image will be drawn within these axes. projection : cartopy.crs.Projection, optional The map projection to be used. If both `ax` and `projection` are specified, this will be checked against the `projection` attribute of `ax`, if it exists. **kwargs : dict, optional Any further keyword arguments are passed to `matplotlib.pyplot.imshow` or `matplotlib.axes.Axes.imshow`, where appropriate. Returns ------- matplotlib.image.AxesImage Raises ------ ValueError If `ax` and `projection` are both specified, but do not match (i.e. `ax.projection != projection`). """ for kw in ("origin", "extent"): if kw in kwargs.keys(): raise TypeError( "imshow got an unexpected keyword argument: {}".format(kw) ) if ax is None: existing_figure = len(plt.get_fignums()) > 0 current_axes = plt.gca() if projection is None: ax = current_axes elif ( isinstance(current_axes, _GeoAxes) and current_axes.projection == projection ): ax = current_axes else: if not existing_figure: current_axes.remove() ax = plt.axes(projection=projection) elif projection is not None: # projection and ax both specified if isinstance(ax, _GeoAxes) and ax.projection == projection: pass # projections match else: raise ValueError( "Both `projection` and `ax` were specified, but" + " `projection` does not match `ax.projection`" ) if isinstance(ax, _GeoAxes) and "transform" not in kwargs.keys(): kwargs["transform"] = _PlateCarree() extent = self.extent if self.origin == "upper": extent = ( extent[0], extent[1], extent[3], extent[2], ) im = ax.imshow(self.data, origin=self.origin, extent=extent, **kwargs) return im def reconstruct(self, time, fill_value=None, partitioning_features=None, threads=1, anchor_plate_id=0, inplace=False, return_array=False)-
Reconstruct raster data to a given time.
Parameters
time:float- Time to which the data will be reconstructed.
fill_value:float, int, str,ortuple, optional- The value to be used for regions outside of the static polygons
at
time. By default (fill_value=None), this value will be determined based on the input. partitioning_features:sequenceofFeatureorstr, optional- The features used to partition the raster grid and assign plate
IDs. By default,
self.plate_reconstruction.static_polygonswill be used, but alternatively any valid argument topygplates.FeaturesFunctionArgumentcan be specified here. threads:int, default1- Number of threads to use for certain computationally heavy routines.
anchor_plate_id:int, default0- ID of the anchored plate.
inplace:bool, defaultFalse- Perform the reconstruction in-place (replace the raster's data with the reconstructed data).
return_array:bool, defaultFalse- Return a
numpy.ndarray, rather than aRaster.
Returns
Rasterornp.ndarray- The reconstructed grid. Areas for which no plate ID could be
determined will be filled with
fill_value.
Raises
TypeError- If this
Rasterhas noplate_reconstructionset.
Notes
For two-dimensional grids,
fill_valueshould be a single number. The default value will benp.nanfor float or complex types, the minimum value for integer types, and the maximum value for unsigned types. For RGB image grids,fill_valueshould be a 3-tuple RGB colour code or a matplotlib colour string. The default value will be black (0.0, 0.0, 0.0) or (0, 0, 0). For RGBA image grids,fill_valueshould be a 4-tuple RGBA colour code or a matplotlib colour string. The default fill value will be transparent black (0.0, 0.0, 0.0, 0.0) or (0, 0, 0, 0).Expand source code
def reconstruct( self, time, fill_value=None, partitioning_features=None, threads=1, anchor_plate_id=0, inplace=False, return_array=False, ): """Reconstruct raster data to a given time. Parameters ---------- time : float Time to which the data will be reconstructed. fill_value : float, int, str, or tuple, optional The value to be used for regions outside of the static polygons at `time`. By default (`fill_value=None`), this value will be determined based on the input. partitioning_features : sequence of Feature or str, optional The features used to partition the raster grid and assign plate IDs. By default, `self.plate_reconstruction.static_polygons` will be used, but alternatively any valid argument to `pygplates.FeaturesFunctionArgument` can be specified here. threads : int, default 1 Number of threads to use for certain computationally heavy routines. anchor_plate_id : int, default 0 ID of the anchored plate. inplace : bool, default False Perform the reconstruction in-place (replace the raster's data with the reconstructed data). return_array : bool, default False Return a `numpy.ndarray`, rather than a `Raster`. Returns ------- Raster or np.ndarray The reconstructed grid. Areas for which no plate ID could be determined will be filled with `fill_value`. Raises ------ TypeError If this `Raster` has no `plate_reconstruction` set. Notes ----- For two-dimensional grids, `fill_value` should be a single number. The default value will be `np.nan` for float or complex types, the minimum value for integer types, and the maximum value for unsigned types. For RGB image grids, `fill_value` should be a 3-tuple RGB colour code or a matplotlib colour string. The default value will be black (0.0, 0.0, 0.0) or (0, 0, 0). For RGBA image grids, `fill_value` should be a 4-tuple RGBA colour code or a matplotlib colour string. The default fill value will be transparent black (0.0, 0.0, 0.0, 0.0) or (0, 0, 0, 0). """ if time < 0.0: raise ValueError("Invalid time: {}".format(time)) time = float(time) if self.plate_reconstruction is None: raise TypeError( "Cannot perform reconstruction - " + "`plate_reconstruction` has not been set" ) if partitioning_features is None: partitioning_features = self.plate_reconstruction.static_polygons result = reconstruct_grid( grid=self.data, partitioning_features=partitioning_features, rotation_model=self.plate_reconstruction.rotation_model, from_time=self.time, to_time=time, extent=self.extent, origin=self.origin, fill_value=fill_value, threads=threads, anchor_plate_id=anchor_plate_id, ) if inplace: self.data = result self._time = time if return_array: return result return self if not return_array: result = type(self)( data=result, plate_reconstruction=self.plate_reconstruction, extent=self.extent, time=time, origin=self.origin, ) return result def resample(self, spacingX, spacingY, method='linear', inplace=False)-
Resample the
gridpassed to theRasterobject with a newspacingXandspacingYusing linear interpolation.Notes
Ultimately,
resamplechanges the lat-lon resolution of the gridded data. The larger the x and y spacings given are, the larger the pixellation of raster data.resamplecreates new latitude and longitude arrays with specified spacings in the X and Y directions (spacingXandspacingY). These arrays are linearly interpolated into a new raster. Ifinplaceis set toTrue, the respaced latitude array, longitude array and raster will inplace the ones currently attributed to theRasterobject.Parameters
spacingX,spacingY:ndarray- Specify the spacing in the X and Y directions with which to resample. The larger
spacingXandspacingYare, the larger the raster pixels become (less resolved). Note: to keep the size of the raster consistent, setspacingX = spacingY; otherwise, if for examplespacingX > spacingY, the raster will appear stretched longitudinally. method:strorint; default: 'linear'- The order of spline interpolation. Must be an integer in the range 0-5. 'nearest', 'linear', and 'cubic' are aliases for 0, 1, and 3, respectively.
inplace:bool, default=False- Choose to overwrite the data (the
self.dataattribute), latitude array (self.lats) and longitude array (self.lons) currently attributed to theRasterobject.
Returns
Raster- The resampled grid. If
inplaceis set toTrue, this raster overwrites the one attributed todata.
Expand source code
def resample(self, spacingX, spacingY, method="linear", inplace=False): """Resample the `grid` passed to the `Raster` object with a new `spacingX` and `spacingY` using linear interpolation. Notes ----- Ultimately, `resample` changes the lat-lon resolution of the gridded data. The larger the x and y spacings given are, the larger the pixellation of raster data. `resample` creates new latitude and longitude arrays with specified spacings in the X and Y directions (`spacingX` and `spacingY`). These arrays are linearly interpolated into a new raster. If `inplace` is set to `True`, the respaced latitude array, longitude array and raster will inplace the ones currently attributed to the `Raster` object. Parameters ---------- spacingX, spacingY : ndarray Specify the spacing in the X and Y directions with which to resample. The larger `spacingX` and `spacingY` are, the larger the raster pixels become (less resolved). Note: to keep the size of the raster consistent, set `spacingX = spacingY`; otherwise, if for example `spacingX > spacingY`, the raster will appear stretched longitudinally. method : str or int; default: 'linear' The order of spline interpolation. Must be an integer in the range 0-5. 'nearest', 'linear', and 'cubic' are aliases for 0, 1, and 3, respectively. inplace : bool, default=False Choose to overwrite the data (the `self.data` attribute), latitude array (`self.lats`) and longitude array (`self.lons`) currently attributed to the `Raster` object. Returns ------- Raster The resampled grid. If `inplace` is set to `True`, this raster overwrites the one attributed to `data`. """ spacingX = np.abs(spacingX) spacingY = np.abs(spacingY) if self.origin == "upper": spacingY *= -1.0 lons = np.arange(self.extent[0], self.extent[1]+spacingX, spacingX) lats = np.arange(self.extent[2], self.extent[3]+spacingY, spacingY) lonq, latq = np.meshgrid(lons, lats) data = self.interpolate(lonq, latq, method=method) if inplace: self._data = data self._lons = lons self._lats = lats else: return Raster(data, self.plate_reconstruction, self.extent, self.time) def resize(self, resX, resY, inplace=False, method='linear', return_array=False)-
Resize the grid passed to the
Rasterobject with a new x and y resolution (resXandresY) using linear interpolation.Notes
Ultimately,
resize"stretches" a raster in the x and y directions. The larger the resolutions in x and y, the more stretched the raster appears in x and y.It creates new latitude and longitude arrays with specific resolutions in the X and Y directions (
resXandresY). These arrays are linearly interpolated into a new raster. Ifinplaceis set toTrue, the resized latitude, longitude arrays and raster will inplace the ones currently attributed to theRasterobject.Parameters
resX,resY:ndarray- Specify the resolutions with which to resize the raster. The larger
resXis, the more longitudinally-stretched the raster becomes. The largerresYis, the more latitudinally-stretched the raster becomes. method:strorint; default: 'linear'- The order of spline interpolation. Must be an integer in the range 0-5. 'nearest', 'linear', and 'cubic' are aliases for 0, 1, and 3, respectively.
inplace:bool, default=False- Choose to overwrite the data (the
self.dataattribute), latitude array (self.lats) and longitude array (self.lons) currently attributed to theRasterobject. return_array:bool, defaultFalse- Return a
numpy.ndarray, rather than aRaster.
Returns
Raster- The resized grid. If
inplaceis set toTrue, this raster overwrites the one attributed todata.
Expand source code
def resize(self, resX, resY, inplace=False, method="linear", return_array=False): """Resize the grid passed to the `Raster` object with a new x and y resolution (`resX` and `resY`) using linear interpolation. Notes ----- Ultimately, `resize` "stretches" a raster in the x and y directions. The larger the resolutions in x and y, the more stretched the raster appears in x and y. It creates new latitude and longitude arrays with specific resolutions in the X and Y directions (`resX` and `resY`). These arrays are linearly interpolated into a new raster. If `inplace` is set to `True`, the resized latitude, longitude arrays and raster will inplace the ones currently attributed to the `Raster` object. Parameters ---------- resX, resY : ndarray Specify the resolutions with which to resize the raster. The larger `resX` is, the more longitudinally-stretched the raster becomes. The larger `resY` is, the more latitudinally-stretched the raster becomes. method : str or int; default: 'linear' The order of spline interpolation. Must be an integer in the range 0-5. 'nearest', 'linear', and 'cubic' are aliases for 0, 1, and 3, respectively. inplace : bool, default=False Choose to overwrite the data (the `self.data` attribute), latitude array (`self.lats`) and longitude array (`self.lons`) currently attributed to the `Raster` object. return_array : bool, default False Return a `numpy.ndarray`, rather than a `Raster`. Returns ------- Raster The resized grid. If `inplace` is set to `True`, this raster overwrites the one attributed to `data`. """ # construct grid lons = np.linspace(self.extent[0], self.extent[1], resX) lats = np.linspace(self.extent[2], self.extent[3], resY) lonq, latq = np.meshgrid(lons, lats) data = self.interpolate(lonq, latq, method=method) if inplace: self._data = data self._lons = lons self._lats = lats if return_array: return data else: return Raster(data, self.plate_reconstruction, self.extent, self.time) def rotate_reference_frames(self, grid_spacing_degrees, reconstruction_time, from_rotation_features_or_model, to_rotation_features_or_model, from_rotation_reference_plate=0, to_rotation_reference_plate=0, non_reference_plate=701, output_name=None)-
Expand source code
def rotate_reference_frames( self, grid_spacing_degrees, reconstruction_time, from_rotation_features_or_model, # filename(s), or pyGPlates feature(s)/collection(s) or a RotationModel to_rotation_features_or_model, # filename(s), or pyGPlates feature(s)/collection(s) or a RotationModel from_rotation_reference_plate=0, to_rotation_reference_plate=0, non_reference_plate=701, output_name=None ): import time as timer """Rotate a grid defined in one plate model reference frame within a gplately.Raster object to another plate reconstruction model reference frame. Parameters ---------- grid_spacing_degrees : float The spacing (in degrees) for the output rotated grid. reconstruction_time : float The time at which to rotate the input grid. from_rotation_features_or_model : str, list of str, or instance of pygplates.RotationModel A filename, or a list of filenames, or a pyGPlates RotationModel object that defines the rotation model that the input grid is currently associated with. to_rotation_features_or_model : str, list of str, or instance of pygplates.RotationModel A filename, or a list of filenames, or a pyGPlates RotationModel object that defines the rotation model that the input grid shall be rotated with. from_rotation_reference_plate : int, default = 0 The current reference plate for the plate model the grid is defined in. Defaults to the anchor plate 0. to_rotation_reference_plate : int, default = 0 The desired reference plate for the plate model the grid is being rotated to. Defaults to the anchor plate 0. non_reference_plate : int, default = 701 An arbitrary placeholder reference frame with which to define the "from" and "to" reference frames. output_name : str, default None If passed, the rotated grid is saved as a netCDF grid to this filename. Returns ------- gplately.Raster() An instance of the gplately.Raster object containing the rotated grid. """ input_positions = [] # Create the pygplates.FiniteRotation that rotates # between the two reference frames. from_rotation_model = pygplates.RotationModel( from_rotation_features_or_model ) to_rotation_model = pygplates.RotationModel( to_rotation_features_or_model ) from_rotation = from_rotation_model.get_rotation( reconstruction_time, non_reference_plate, anchor_plate_id=from_rotation_reference_plate ) to_rotation = to_rotation_model.get_rotation( reconstruction_time, non_reference_plate, anchor_plate_id=to_rotation_reference_plate ) reference_frame_conversion_rotation = to_rotation * from_rotation.get_inverse() # Resize the input grid to the specified output resolution before rotating resX = _deg2pixels( grid_spacing_degrees, self.extent[0], self.extent[1] ) resY = _deg2pixels( grid_spacing_degrees, self.extent[2], self.extent[3] ) resized_input_grid = self.resize( resX, resY, inplace=False ) # Get the flattened lons, lats llons, llats = np.meshgrid( resized_input_grid.lons, resized_input_grid.lats ) llons = llons.flatten() llats = llats.flatten() input_coords = [(llons[i], llats[i]) for i in range(0, len(llons))] # Convert lon-lat points of Raster grid to pyGPlates points input_points = pygplates.MultiPointOnSphere( (lat, lon) for lon, lat in input_coords ) # Get grid values of the resized Raster object values = np.array(resized_input_grid.data).flatten() # Rotate grid nodes to the other reference frame output_points = reference_frame_conversion_rotation * input_points # Assemble rotated points with grid values. out_lon = [] out_lat = [] zdata = [] for index, point in enumerate(output_points): lat, lon = point.to_lat_lon() out_lon.append(lon) out_lat.append(lat) zdata.append(values[index]) # Create a regular grid on which to interpolate lats, lons and zdata # Use the extent of the original Raster object extent_globe = self.extent resX = int(np.floor((extent_globe[1] - extent_globe[0]) / grid_spacing_degrees)) + 1 resY = int(np.floor((extent_globe[3] - extent_globe[2]) / grid_spacing_degrees)) + 1 grid_lon = np.linspace( extent_globe[0], extent_globe[1], resX ) grid_lat = np.linspace( extent_globe[2], extent_globe[3], resY ) X, Y = np.meshgrid( grid_lon, grid_lat ) # Interpolate lons, lats and zvals over a regular grid using nearest # neighbour interpolation Z = griddata( (out_lon, out_lat), zdata, (X, Y), method='nearest' ) # Write output grid to netCDF if requested. if output_name: write_netcdf_grid( output_name, Z, extent=extent_globe, ) return Raster(data=Z) def save_to_netcdf4(self, filename)-
Saves the grid attributed to the
Rasterobject to the givenfilename(including the ".nc" extension) in netCDF4 format.Expand source code
def save_to_netcdf4(self, filename): """ Saves the grid attributed to the `Raster` object to the given `filename` (including the ".nc" extension) in netCDF4 format.""" write_netcdf_grid(str(filename), self.data, self.extent)
class SeafloorGrid (PlateReconstruction_object, PlotTopologies_object, max_time, min_time, ridge_time_step, save_directory='agegrids', file_collection=None, refinement_levels=5, ridge_sampling=0.5, extent=(-180, 180, -90, 90), grid_spacing=None, subduction_collision_parameters=(5.0, 10.0), initial_ocean_mean_spreading_rate=75.0, resume_from_checkpoints=False, zval_names=('SPREADING_RATE',), continent_mask_filename=None)-
A class to generate grids that track data atop global ocean basin points (which emerge from mid ocean ridges) through geological time.
Parameters
PlateReconstruction_object:instanceof<gplately.PlateReconstruction>- A GPlately PlateReconstruction object with a
and a containing topology features. PlotTopologies_object:instanceof<gplately.PlotTopologies>- A GPlately PlotTopologies object with a continental polygon or COB terrane polygon file to mask grids with.
max_time:float- The maximum time for age gridding.
min_time:float- The minimum time for age gridding.
ridge_time_step:float- The delta time for resolving ridges (and thus age gridding).
save_directory:str, defaultNone'- The top-level directory to save all outputs to.
file_collection:str, defaultNone- A string to identify the plate model used (will be automated later).
refinement_levels:int, default5- Control the number of points in the icosahedral mesh (higher integer means higher resolution of continent masks).
ridge_sampling:float, default0.5- Spatial resolution (in degrees) at which points that emerge from ridges are tessellated.
extent:listoffloatorint, default[-180.,180.,-90.,90.]- A list containing the mininum longitude, maximum longitude, minimum latitude and maximum latitude extents for all masking and final grids.
grid_spacing:float, defaultNone- The degree spacing/interval with which to space grid points across all masking and
final grids. If
grid_spacingis provided, all grids will use it. If not,grid_spacingdefaults to 0.1. subduction_collision_parameters:len-2 tupleoffloat, default(5.0, 10.0)- A 2-tuple of (threshold velocity delta in kms/my, threshold distance to boundary per My in kms/my)
- initial_ocean_mean_spreading_rate : float, default 75.
- A spreading rate to uniformly allocate to points that define the initial ocean
- basin. These points will have inaccurate ages, but most of them will be phased
- out after points with plate-model prescribed ages emerge from ridges and spread
- to push them towards collision boundaries (where they are deleted).
resume_from_checkpoints:bool, defaultFalse- If set to
True, and the gridding preparation stage (continental masking and/or ridge seed building) is interrupted, SeafloorGrids will resume gridding preparation from the last successful preparation time. If set toFalse, SeafloorGrids will automatically overwrite all files insave_directoryif re-run after interruption, or normally re-run, thus beginning gridding preparation from scratch.Falsewill be useful if data allocated to the MOR seed points need to be augmented. zval_names:listofstr- A list containing string labels for the z values to attribute to points. Will be used as column headers for z value point dataframes.
continent_mask_filename:str- An optional parameter pointing to the full path to a continental mask for each timestep. Assuming the time is in the filename, i.e. "/path/to/continent_mask_0Ma.nc", it should be passed as "/path/to/continent_mask_{}Ma.nc" with curly brackets. Include decimal formatting if needed.
Expand source code
class SeafloorGrid(object): """A class to generate grids that track data atop global ocean basin points (which emerge from mid ocean ridges) through geological time. Parameters ---------- PlateReconstruction_object : instance of <gplately.PlateReconstruction> A GPlately PlateReconstruction object with a <pygplates.RotationModel> and a <pygplates.FeatureCollection> containing topology features. PlotTopologies_object : instance of <gplately.PlotTopologies> A GPlately PlotTopologies object with a continental polygon or COB terrane polygon file to mask grids with. max_time : float The maximum time for age gridding. min_time : float The minimum time for age gridding. ridge_time_step : float The delta time for resolving ridges (and thus age gridding). save_directory : str, default None' The top-level directory to save all outputs to. file_collection : str, default None A string to identify the plate model used (will be automated later). refinement_levels : int, default 5 Control the number of points in the icosahedral mesh (higher integer means higher resolution of continent masks). ridge_sampling : float, default 0.5 Spatial resolution (in degrees) at which points that emerge from ridges are tessellated. extent : list of float or int, default [-180.,180.,-90.,90.] A list containing the mininum longitude, maximum longitude, minimum latitude and maximum latitude extents for all masking and final grids. grid_spacing : float, default None The degree spacing/interval with which to space grid points across all masking and final grids. If `grid_spacing` is provided, all grids will use it. If not, `grid_spacing` defaults to 0.1. subduction_collision_parameters : len-2 tuple of float, default (5.0, 10.0) A 2-tuple of (threshold velocity delta in kms/my, threshold distance to boundary per My in kms/my) initial_ocean_mean_spreading_rate : float, default 75. A spreading rate to uniformly allocate to points that define the initial ocean basin. These points will have inaccurate ages, but most of them will be phased out after points with plate-model prescribed ages emerge from ridges and spread to push them towards collision boundaries (where they are deleted). resume_from_checkpoints : bool, default False If set to `True`, and the gridding preparation stage (continental masking and/or ridge seed building) is interrupted, SeafloorGrids will resume gridding preparation from the last successful preparation time. If set to `False`, SeafloorGrids will automatically overwrite all files in `save_directory` if re-run after interruption, or normally re-run, thus beginning gridding preparation from scratch. `False` will be useful if data allocated to the MOR seed points need to be augmented. zval_names : list of str A list containing string labels for the z values to attribute to points. Will be used as column headers for z value point dataframes. continent_mask_filename : str An optional parameter pointing to the full path to a continental mask for each timestep. Assuming the time is in the filename, i.e. "/path/to/continent_mask_0Ma.nc", it should be passed as "/path/to/continent_mask_{}Ma.nc" with curly brackets. Include decimal formatting if needed. """ def __init__( self, PlateReconstruction_object, PlotTopologies_object, max_time, min_time, ridge_time_step, save_directory="agegrids", file_collection=None, refinement_levels=5, ridge_sampling=0.5, extent=(-180, 180, -90, 90), grid_spacing=None, subduction_collision_parameters=(5.0, 10.0), initial_ocean_mean_spreading_rate=75.0, resume_from_checkpoints=False, zval_names=("SPREADING_RATE",), continent_mask_filename=None, ): # Provides a rotation model, topology features and reconstruction time for # the SeafloorGrid self.PlateReconstruction_object = PlateReconstruction_object self.rotation_model = self.PlateReconstruction_object.rotation_model self.topology_features = self.PlateReconstruction_object.topology_features self._PlotTopologies_object = PlotTopologies_object save_directory = str(save_directory) if not os.path.isdir(save_directory): print("Output directory does not exist; creating now: " + save_directory) os.makedirs(save_directory, exist_ok=True) self.save_directory = save_directory if file_collection is not None: file_collection = str(file_collection) self.file_collection = file_collection # Topological parameters self.refinement_levels = refinement_levels self.ridge_sampling = ridge_sampling self.subduction_collision_parameters = subduction_collision_parameters self.initial_ocean_mean_spreading_rate = initial_ocean_mean_spreading_rate # Gridding parameters self.extent = extent # A list of degree spacings that allow an even division of the global lat-lon extent. divisible_degree_spacings = [0.1, 0.25, 0.5, 0.75, 1.0] if grid_spacing: # If the provided degree spacing is in the list of permissible spacings, use it # and prepare the number of pixels in x and y (spacingX and spacingY) if grid_spacing in divisible_degree_spacings: self.grid_spacing = grid_spacing self.spacingX = _deg2pixels( grid_spacing, self.extent[0], self.extent[1] ) self.spacingY = _deg2pixels( grid_spacing, self.extent[2], self.extent[3] ) # If the provided spacing is >>1 degree, use 1 degree elif grid_spacing >= divisible_degree_spacings[-1]: self.grid_spacing = divisible_degree_spacings[-1] self.spacingX = _deg2pixels( divisible_degree_spacings[-1], self.extent[0], self.extent[1] ) self.spacingY = _deg2pixels( divisible_degree_spacings[-1], self.extent[2], self.extent[3] ) with warnings.catch_warnings(): warnings.simplefilter("always") warnings.warn( "The provided grid_spacing of {} is quite large. To preserve the grid resolution, a {} degree spacing has been employed instead".format( grid_spacing, self.grid_spacing ) ) # If the provided degree spacing is not in the list of permissible spacings, but below # a degree, find the closest permissible degree spacing. Use this and find # spacingX and spacingY. else: for divisible_degree_spacing in divisible_degree_spacings: # The tolerance is half the difference between consecutive divisible spacings. # Max is 1 degree for now - other integers work but may provide too little of a # grid resolution. if abs(grid_spacing - divisible_degree_spacing) <= 0.125: new_deg_res = divisible_degree_spacing self.grid_spacing = new_deg_res self.spacingX = _deg2pixels( new_deg_res, self.extent[0], self.extent[1] ) self.spacingY = _deg2pixels( new_deg_res, self.extent[2], self.extent[3] ) with warnings.catch_warnings(): warnings.simplefilter("always") warnings.warn( "The provided grid_spacing of {} does not cleanly divide into the global extent. A degree spacing of {} has been employed instead.".format( grid_spacing, self.grid_spacing ) ) else: # If a spacing degree is not provided, use default # resolution and get default spacingX and spacingY self.grid_spacing = 0.1 self.spacingX = 3601 self.spacingY = 1801 self.resume_from_checkpoints = resume_from_checkpoints # Temporal parameters self._max_time = float(max_time) self.min_time = float(min_time) self.ridge_time_step = float(ridge_time_step) self.time_array = np.arange( self._max_time, self.min_time - 0.1, -self.ridge_time_step ) # If PlotTopologies' time attribute is not equal to the maximum time in the # seafloor grid reconstruction tree, make it equal. This will ensure the time # for continental masking is consistent. if self._PlotTopologies_object.time != self._max_time: self._PlotTopologies_object.time = self._max_time # Essential features and meshes for the SeafloorGrid self.continental_polygons = ensure_polygon_geometry( self._PlotTopologies_object.continents, self.rotation_model, self._max_time ) self._PlotTopologies_object.continents = PlotTopologies_object.continents ( self.icosahedral_multi_point, self.icosahedral_global_mesh, ) = create_icosahedral_mesh(self.refinement_levels) # Z value parameters self.zval_names = zval_names self.default_column_headers = [ "CURRENT_LONGITUDES", "CURRENT_LATITUDES", "SEAFLOOR_AGE", "BIRTH_LAT_SNAPSHOT", "POINT_ID_SNAPSHOT", ] self.total_column_headers = np.concatenate( [self.default_column_headers, self.zval_names] ) # Filename for continental masks that the user can provide instead of building it here self.continent_mask_filename = continent_mask_filename # If the user provides a continental mask filename, we need to downsize the mask # resolution for when we create the initial ocean mesh. The mesh does not need to be high-res. if self.continent_mask_filename is not None: # Determine which percentage to use to scale the continent mask resolution at max time def _map_res_to_node_percentage(self, continent_mask_filename): maskY, maskX = grids.read_netcdf_grid( continent_mask_filename.format(self._max_time) ).shape mask_deg = _pixels2deg(maskX, self.extent[0], self.extent[1]) if mask_deg <= 0.1: percentage = 0.1 elif mask_deg <= 0.25: percentage = 0.3 elif mask_deg <= 0.5: percentage = 0.5 elif mask_deg < 0.75: percentage = 0.6 elif mask_deg >= 1: percentage = 0.75 return mask_deg, percentage _, self.percentage = _map_res_to_node_percentage( self, self.continent_mask_filename ) # Allow SeafloorGrid time to be updated, and to update the internally-used # PlotTopologies' time attribute too. If PlotTopologies is used outside the # object, its `time` attribute is not updated. @property def max_time(self): """The reconstruction time.""" return self._max_time @property def PlotTopologiesTime(self): return self._PlotTopologies_object.time @max_time.setter def max_time(self, var): if var >= 0: self.update_time(var) else: raise ValueError("Enter a valid time >= 0") def update_time(self, max_time): self._max_time = float(max_time) self._PlotTopologies_object.time = float(max_time) def _collect_point_data_in_dataframe( self, pygplates_featurecollection, zval_ndarray, time ): """At a given timestep, create a pandas dataframe holding all attributes of point features. Rather than store z values as shapefile attributes, store them in a dataframe indexed by feature ID. """ # Turn the zval_ndarray into a numPy array zval_ndarray = np.array(zval_ndarray) feature_id = [] for feature in pygplates_featurecollection: feature_id.append(str(feature.get_feature_id())) # Prepare the zval ndarray (can be of any shape) to be saved with default point data zvals_to_store = {} # If only one zvalue (fow now, spreading rate) if zval_ndarray.ndim == 1: zvals_to_store[self.zval_names[0]] = zval_ndarray data_to_store = [zvals_to_store[i] for i in zvals_to_store] else: for i in zval_ndarray.shape[1]: zvals_to_store[self.zval_names[i]] = [ list(j) for j in zip(*zval_ndarray) ][i] data_to_store = [zvals_to_store[i] for i in zvals_to_store] basename = "point_data_dataframe_{}Ma".format(time) if self.file_collection is not None: basename = "{}_{}".format(self.file_collection, basename) filename = os.path.join(self.save_directory, basename) np.savez_compressed(filename, FEATURE_ID=feature_id, *data_to_store) return def create_initial_ocean_seed_points(self): """Create the initial ocean basin seed point domain (at `max_time` only) using Stripy's icosahedral triangulation with the specified `self.refinement_levels`. The ocean mesh starts off as a global-spanning Stripy icosahedral mesh. `create_initial_ocean_seed_points` passes the automatically-resolved-to-current-time continental polygons from the `PlotTopologies_object`'s `continents` attribute (which can be from a COB terrane file or a continental polygon file) into Plate Tectonic Tools' point-in-polygon routine. It identifies ocean basin points that lie: * outside the polygons (for the ocean basin point domain) * inside the polygons (for the continental mask) Points from the mesh outside the continental polygons make up the ocean basin seed point mesh. The masked mesh is outputted as a compressed GPML (GPMLZ) file with the filename: "ocean_basin_seed_points_{}Ma.gpmlz" if a `save_directory` is passed. Otherwise, the mesh is returned as a pyGPlates FeatureCollection object. Notes ----- This point mesh represents ocean basin seafloor that was produced before `SeafloorGrid.max_time`, and thus has unknown properties like valid time and spreading rate. As time passes, the plate reconstruction model sees points emerging from MORs. These new points spread to occupy the ocean basins, moving the initial filler points closer to subduction zones and continental polygons with which they can collide. If a collision is detected by `PlateReconstruction`s `ReconstructByTopologies` object, these points are deleted. Ideally, if a reconstruction tree spans a large time range, **all** initial mesh points would collide with a continent or be subducted, leaving behind a mesh of well-defined MOR-emerged ocean basin points that data can be attributed to. However, some of these initial points situated close to contiental boundaries are retained through time - these form point artefacts with anomalously high ages. Even deep-time plate models (e.g. 1 Ga) will have these artefacts - removing them would require more detail to be added to the reconstruction model. Returns ------- ocean_basin_point_mesh : pygplates.FeatureCollection of pygplates.MultiPointOnSphere A feature collection of point objects on the ocean basin. """ if self.continent_mask_filename is None: # Ensure COB terranes at max time have reconstruction IDs and valid times COB_polygons = ensure_polygon_geometry( self._PlotTopologies_object.continents, self.rotation_model, self._max_time, ) # zval is a binary array encoding whether a point # coordinate is within a COB terrane polygon or not. # Use the icosahedral mesh MultiPointOnSphere attribute _, ocean_basin_point_mesh, zvals = point_in_polygon_routine( self.icosahedral_multi_point, COB_polygons ) # Plates to partition with plate_partitioner = pygplates.PlatePartitioner( COB_polygons, self.rotation_model, ) # Plate partition the ocean basin points meshnode_feature = pygplates.Feature( pygplates.FeatureType.create_from_qualified_string("gpml:MeshNode") ) meshnode_feature.set_geometry( ocean_basin_point_mesh # multi_point ) ocean_basin_meshnode = pygplates.FeatureCollection(meshnode_feature) paleogeography = plate_partitioner.partition_features( ocean_basin_meshnode, partition_return=pygplates.PartitionReturn.separate_partitioned_and_unpartitioned, properties_to_copy=[pygplates.PropertyName.gpml_shapefile_attributes], ) ocean_points = paleogeography[1] # Separate those inside polygons continent_points = paleogeography[0] # Separate those outside polygons # If a set of continent masks was passed, we can use max_time's continental # mask to build the initial profile of seafloor age. else: max_time_cont_mask = grids.Raster( self.continent_mask_filename.format(self._max_time) ) # If the input grid is at 0.5 degree uniform spacing, then the input # grid is 7x more populated than a 6-level stripy icosahedral mesh and # using this resolution for the initial ocean mesh will dramatically slow down # reconstruction by topologies. # Scale down the resolution based on the input mask resolution # (percentage was found in __init__.) max_time_cont_mask.resize( int(max_time_cont_mask.shape[0] * self.percentage), int(max_time_cont_mask.shape[1] * self.percentage), inplace=True, ) lat = np.linspace(-90, 90, max_time_cont_mask.shape[0]) lon = np.linspace(-180, 180, max_time_cont_mask.shape[1]) llon, llat = np.meshgrid(lon, lat) mask_inds = np.where(max_time_cont_mask.data.flatten() == 0) mask_vals = max_time_cont_mask.data.flatten() mask_lon = llon.flatten()[mask_inds] mask_lat = llat.flatten()[mask_inds] ocean_pt_feature = pygplates.Feature() ocean_pt_feature.set_geometry( pygplates.MultiPointOnSphere(zip(mask_lat, mask_lon)) ) ocean_points = [ocean_pt_feature] # Now that we have ocean points... # Determine age of ocean basin points using their proximity to MOR features # and an assumed globally-uniform ocean basin mean spreading rate. # We need resolved topologies at the `max_time` to pass into the proximity # function resolved_topologies = [] shared_boundary_sections = [] pygplates.resolve_topologies( self.topology_features, self.rotation_model, resolved_topologies, self._max_time, shared_boundary_sections, ) pX, pY, pZ = tools.find_distance_to_nearest_ridge( resolved_topologies, shared_boundary_sections, ocean_points, ) # Divide spreading rate by 2 to use half the mean spreading rate pAge = np.array(pZ) / (self.initial_ocean_mean_spreading_rate / 2.0) initial_ocean_point_features = [] initial_ocean_multipoints = [] for point in zip(pX, pY, pAge): point_feature = pygplates.Feature() point_feature.set_geometry(pygplates.PointOnSphere(point[1], point[0])) # Add 'time' to the age at the time of computation, to get the valid time in Ma point_feature.set_valid_time(point[2] + self._max_time, -1) # For now: custom zvals are added as shapefile attributes - will attempt pandas data frames # point_feature = set_shapefile_attribute(point_feature, self.initial_ocean_mean_spreading_rate, "SPREADING_RATE") # Seems like static data initial_ocean_point_features.append(point_feature) initial_ocean_multipoints.append(point_feature.get_geometry()) # print(initial_ocean_point_features) multi_point_feature = pygplates.MultiPointOnSphere(initial_ocean_multipoints) basename = "ocean_basin_seed_points_{}_RLs_{}Ma.gpmlz".format( self.refinement_levels, self._max_time, ) if self.file_collection is not None: basename = "{}_{}".format(self.file_collection, basename) output_filename = os.path.join(self.save_directory, basename) initial_ocean_feature_collection = pygplates.FeatureCollection( initial_ocean_point_features ) initial_ocean_feature_collection.write(output_filename) # Collect all point feature data into a pandas dataframe self._collect_point_data_in_dataframe( initial_ocean_feature_collection, np.array( [self.initial_ocean_mean_spreading_rate] * len(pX) ), # for now, spreading rate is one zvalue for initial ocean points. will other zvalues need to have a generalised workflow? self._max_time, ) return ( pygplates.FeatureCollection(initial_ocean_point_features), multi_point_feature, ) def _get_mid_ocean_ridge_seedpoints(self, time_array): # Topology features from `PlotTopologies`. topology_features_extracted = pygplates.FeaturesFunctionArgument( self.topology_features ) # Create a mask for each timestep if time_array[0] != self._max_time: print( "MOR seed point building interrupted - resuming at {} Ma!".format( time_array[0] ) ) for time in time_array: # Points and their z values that emerge from MORs at this time. shifted_mor_points = [] point_spreading_rates = [] # Resolve topologies to the current time. resolved_topologies = [] shared_boundary_sections = [] pygplates.resolve_topologies( topology_features_extracted.get_features(), self.rotation_model, resolved_topologies, time, shared_boundary_sections, ) # pygplates.ResolvedTopologicalSection objects. for shared_boundary_section in shared_boundary_sections: if ( shared_boundary_section.get_feature().get_feature_type() == pygplates.FeatureType.create_gpml("MidOceanRidge") ): spreading_feature = shared_boundary_section.get_feature() # Find the stage rotation of the spreading feature in the # frame of reference of its geometry at the current # reconstruction time (the MOR is currently actively spreading). # The stage pole can then be directly geometrically compared # to the *reconstructed* spreading geometry. stage_rotation = separate_ridge_transform_segments.get_stage_rotation_for_reconstructed_geometry( spreading_feature, self.rotation_model, time ) if not stage_rotation: # Skip current feature - it's not a spreading feature. continue # Get the stage pole of the stage rotation. # Note that the stage rotation is already in frame of # reference of the *reconstructed* geometry at the spreading time. stage_pole, _ = stage_rotation.get_euler_pole_and_angle() # One way rotates left and the other right, but don't know # which - doesn't matter in our example though. rotate_slightly_off_mor_one_way = pygplates.FiniteRotation( stage_pole, np.radians(0.01) ) rotate_slightly_off_mor_opposite_way = ( rotate_slightly_off_mor_one_way.get_inverse() ) subsegment_index = [] # Iterate over the shared sub-segments. for ( shared_sub_segment ) in shared_boundary_section.get_shared_sub_segments(): # Tessellate MOR section. mor_points = pygplates.MultiPointOnSphere( shared_sub_segment.get_resolved_geometry().to_tessellated( np.radians(self.ridge_sampling) ) ) coords = mor_points.to_lat_lon_list() lats = [i[0] for i in coords] lons = [i[1] for i in coords] left_plate = ( shared_boundary_section.get_feature().get_left_plate(None) ) right_plate = ( shared_boundary_section.get_feature().get_right_plate(None) ) if left_plate is not None and right_plate is not None: # Get the spreading rates for all points in this sub segment ( spreading_rates, subsegment_index, ) = tools.calculate_spreading_rates( time=time, lons=lons, lats=lats, left_plates=[left_plate] * len(lons), right_plates=[right_plate] * len(lons), rotation_model=self.rotation_model, delta_time=self.ridge_time_step, ) else: spreading_rates = [np.nan] * len(lons) # Loop through all but the 1st and last points in the current sub segment for point, rate in zip( mor_points.get_points()[1:-1], spreading_rates[1:-1], ): # Add the point "twice" to the main shifted_mor_points list; once for a L-side # spread, another for a R-side spread. Then add the same spreading rate twice # to the list - this therefore assumes spreading rate is symmetric. shifted_mor_points.append( rotate_slightly_off_mor_one_way * point ) shifted_mor_points.append( rotate_slightly_off_mor_opposite_way * point ) point_spreading_rates.extend([rate] * 2) # point_indices.extend(subsegment_index) # Summarising get_isochrons_for_ridge_snapshot; # Write out the ridge point born at 'ridge_time' but their position at 'ridge_time - time_step'. mor_point_features = [] for curr_point in shifted_mor_points: feature = pygplates.Feature() feature.set_geometry(curr_point) feature.set_valid_time(time, -999) # delete - time_step # feature.set_name(str(spreading_rate)) # feature = set_shapefile_attribute(feature, spreading_rate, "SPREADING_RATE") # make spreading rate a shapefile attribute mor_point_features.append(feature) mor_points = pygplates.FeatureCollection(mor_point_features) # Write MOR points at `time` to gpmlz basename = "MOR_plus_one_points_{:0.2f}.gpmlz".format(time) if self.file_collection is not None: basename = "{}_{}".format(self.file_collection, basename) mor_points.write(os.path.join(self.save_directory, basename)) # Make sure the max time dataframe is for the initial ocean points only if time != self._max_time: self._collect_point_data_in_dataframe( mor_points, point_spreading_rates, time ) print("Finished building MOR seedpoints at {} Ma!".format(time)) return def build_all_MOR_seedpoints(self): """Resolve mid-ocean ridges for all times between `min_time` and `max_time`, divide them into points that make up their shared sub-segments. Rotate these points to the left and right of the ridge using their stage rotation so that they spread from the ridge. Z-value allocation to each point is done here. In future, a function (like the spreading rate function) to calculate general z-data will be an input parameter. Notes ----- If MOR seed point building is interrupted, progress is safeguarded as long as `resume_from_checkpoints` is set to `True`. This assumes that points spread from ridges symmetrically, with the exception of large ridge jumps at successive timesteps. Therefore, z-values allocated to ridge-emerging points will appear symmetrical until changes in spreading ridge geometries create asymmetries. In future, this will have a checkpoint save feature so that execution (which occurs during preparation for ReconstructByTopologies and can take several hours) can be safeguarded against run interruptions. Parameters ---------- time : float The time at which to resolve ridge points and stage-rotate them off the ridge. Returns ------- mor_point_features : FeatureCollection All ridge seed points that have emerged from all ridge topologies at `time`. These points have spread by being slightly rotated away from ridge locations at `time`. References ---------- get_mid_ocean_ridge_seedpoints() has been adapted from https://github.com/siwill22/agegrid-0.1/blob/master/automatic_age_grid_seeding.py#L117. """ # If we mustn't overwrite existing files in the `save_directory`, check the status of MOR seeding # to know where to start/continue seeding if self.resume_from_checkpoints: # Check the last MOR seedpoint gpmlz file that was built checkpointed_MOR_seedpoints = [ s.split("/")[-1] for s in glob.glob(self.save_directory + "/" + "*MOR_plus_one_points*") ] try: # -2 as an index accesses the age (float type), safeguards against identifying numbers in the SeafloorGrid.file_collection string last_seed_time = np.sort( [ float(re.findall(r"\d+", s)[-2]) for s in checkpointed_MOR_seedpoints ] )[0] # If none were built yet except: last_seed_time = "nil" # If MOR seeding has not started, start it from the top if last_seed_time == "nil": time_array = self.time_array # If the last seed time it could identify is outside the time bounds of the current instance of SeafloorGrid, start # from the top (this may happen if we use the same save directory for grids for a new set of times) elif last_seed_time not in self.time_array: time_array = self.time_array # If seeding was done to the min_time, we are finished elif last_seed_time == self.min_time: return # If seeding to `min_time` has been interrupted, resume it at last_masked_time. else: time_array = np.arange( last_seed_time, self.min_time - 0.1, -self.ridge_time_step ) # If we must overwrite all files in `save_directory`, start from `max_time`. else: time_array = self.time_array # Build all continental masks and spreading ridge points (with z values) self._get_mid_ocean_ridge_seedpoints(time_array) return def _create_continental_mask(self, time_array): """Create a continental mask for each timestep.""" if time_array[0] != self._max_time: print( "Masking interrupted - resuming continental mask building at {} Ma!".format( time_array[0] ) ) for time in time_array: self._PlotTopologies_object.time = time geoms = self._PlotTopologies_object.continents final_grid = grids.rasterise( geoms, key=1.0, shape=(self.spacingY, self.spacingX), extent=self.extent, origin="lower", ) final_grid[np.isnan(final_grid)] = 0.0 output_basename = "continent_mask_{}Ma.nc".format(time) if self.file_collection is not None: output_basename = "{}_{}".format( self.file_collection, output_basename, ) output_filename = os.path.join( self.save_directory, output_basename, ) grids.write_netcdf_grid( output_filename, final_grid, extent=[-180, 180, -90, 90] ) print("Finished building a continental mask at {} Ma!".format(time)) return def build_all_continental_masks(self): """Create a continental mask to define the ocean basin for all times between `min_time` and `max_time`. as well as to use as continental collision boundaries in `ReconstructByTopologies`. Notes ----- Continental masking progress is safeguarded if ever masking is interrupted, provided that `resume_from_checkpoints` is set to `True`. If `ReconstructByTopologies` identifies a continental collision between oceanic points and the boundaries of this continental mask at `time`, those points are deleted at `time`. The continental mask is also saved to "/continent_mask_{}Ma.nc" as a compressed netCDF4 file if a `save_directory` is passed. Otherwise, the final grid is returned as a NumPy ndarray object. Returns ------- all_continental_masks : list of ndarray A masked grid per timestep in `time_array` with 1=continental point, and 0=ocean point, for all points on the full global icosahedral mesh. """ # If we mustn't overwrite existing files in the `save_directory`, check the status # of continental masking to know where to start/continue masking if self.resume_from_checkpoints: # Check the last continental mask that could be built checkpointed_continental_masks = [ s.split("/")[-1] for s in glob.glob(self.save_directory + "/" + "*continent_mask*") ] try: # -2 as an index accesses the age (float type), safeguards against identifying numbers in the SeafloorGrid.file_collection string last_masked_time = np.sort( [ float(re.findall(r"\d+", s)[-2]) for s in checkpointed_continental_masks ] )[0] # If none were built yet except: last_masked_time = "nil" # If masking has not started, start it from the top if last_masked_time == "nil": time_array = self.time_array # If the last seed time it could identify is outside the time bounds of the current instance of SeafloorGrid, start # from the top (this may happen if we use the same save directory for grids for a new set of times) elif last_masked_time not in self.time_array: time_array = self.time_array # If masking was done to the min_time, we are finished elif last_masked_time == self.min_time: return # If masking to `min_time` has been interrupted, resume it at last_masked_time. else: time_array = np.arange( last_masked_time, self.min_time - 0.1, -self.ridge_time_step ) # If we must overwrite all files in `save_directory`, start from `max_time`. else: time_array = self.time_array # Build all continental masks and spreading ridge points (with z values) self._create_continental_mask(time_array) return def _extract_zvalues_from_npz_to_ndarray(self, featurecollection, time): # NPZ file of seedpoint z values that emerged at this time basename = "point_data_dataframe_{}Ma.npz".format(time) if self.file_collection is not None: basename = "{}_{}".format(self.file_collection, basename) filename = os.path.join(self.save_directory, basename) loaded_npz = np.load(filename) curr_zvalues = np.empty([len(featurecollection), len(self.zval_names)]) for i in range(len(self.zval_names)): # Account for the 0th index being for point feature IDs curr_zvalues[:, i] = np.array(loaded_npz["arr_{}".format(i)]) return curr_zvalues def prepare_for_reconstruction_by_topologies(self): """Prepare three main auxiliary files for seafloor data gridding: * Initial ocean seed points (at `max_time`) * Continental masks (from `max_time` to `min_time`) * MOR points (from `max_time` to `min_time`) Returns lists of all attributes for the initial ocean point mesh and all ridge points for all times in the reconstruction time array. """ # INITIAL OCEAN SEED POINT MESH ---------------------------------------------------- ( initial_ocean_seed_points, initial_ocean_seed_points_mp, ) = self.create_initial_ocean_seed_points() print("Finished building initial_ocean_seed_points!") # MOR SEED POINTS AND CONTINENTAL MASKS -------------------------------------------- # The start time for seeding is controlled by whether the overwrite_existing_gridding_inputs # parameter is set to `True` (in which case the start time is `max_time`). If it is `False` # and; # - a run of seeding and continental masking was interrupted, and ridge points were # checkpointed at n Ma, seeding resumes at n-1 Ma until `min_time` or another interruption # occurs; # - seeding was completed but the subsequent gridding input creation was interrupted, # seeding is assumed completed and skipped. The workflow automatically proceeds to re-gridding. if self.continent_mask_filename is None: self.build_all_continental_masks() else: print( "Continent masks passed to SeafloorGrid - skipping continental mask generation!" ) self.build_all_MOR_seedpoints() # ALL-TIME POINTS ----------------------------------------------------- # Extract all feature attributes for all reconstruction times into lists active_points = [] appearance_time = [] birth_lat = [] # latitude_of_crust_formation prev_lat = [] prev_lon = [] # Extract point feature attributes from MOR seed points all_mor_features = [] zvalues = np.empty((0, len(self.zval_names))) for time in self.time_array: # If we're at the maximum time, start preparing points from the initial ocean mesh # as well as their z values if time == self._max_time: for feature in initial_ocean_seed_points: active_points.append(feature.get_geometry()) appearance_time.append(feature.get_valid_time()[0]) birth_lat.append(feature.get_geometry().to_lat_lon_list()[0][0]) prev_lat.append(feature.get_geometry().to_lat_lon_list()[0][0]) prev_lon.append(feature.get_geometry().to_lat_lon_list()[0][1]) curr_zvalues = self._extract_zvalues_from_npz_to_ndarray( initial_ocean_seed_points, time ) zvalues = np.concatenate((zvalues, curr_zvalues), axis=0) # Otherwise, we'd be preparing MOR points and their z values else: # GPMLZ file of MOR seedpoints basename = "MOR_plus_one_points_{:0.2f}.gpmlz".format(time) if self.file_collection is not None: basename = "{}_{}".format(self.file_collection, basename) filename = os.path.join(self.save_directory, basename) features = pygplates.FeatureCollection(filename) for feature in features: if feature.get_valid_time()[0] < self.time_array[0]: active_points.append(feature.get_geometry()) appearance_time.append(feature.get_valid_time()[0]) birth_lat.append(feature.get_geometry().to_lat_lon_list()[0][0]) prev_lat.append(feature.get_geometry().to_lat_lon_list()[0][0]) prev_lon.append(feature.get_geometry().to_lat_lon_list()[0][1]) # COLLECT NDARRAY OF ALL ZVALUES IN THIS TIMESTEP ------------------ curr_zvalues = self._extract_zvalues_from_npz_to_ndarray(features, time) zvalues = np.concatenate((zvalues, curr_zvalues), axis=0) return active_points, appearance_time, birth_lat, prev_lat, prev_lon, zvalues def reconstruct_by_topologies(self): """Obtain all active ocean seed points at `time` - these are points that have not been consumed at subduction zones or have not collided with continental polygons. All active points' latitudes, longitues, seafloor ages, spreading rates and all other general z-values are saved to a gridding input file (.npz). """ print("Preparing all initial files...") # Obtain all info from the ocean seed points and all MOR points through time, store in # arrays ( active_points, appearance_time, birth_lat, prev_lat, prev_lon, zvalues, ) = self.prepare_for_reconstruction_by_topologies() #### Begin reconstruction by topology process: # Indices for all points (`active_points`) that have existed from `max_time` to `min_time`. point_id = range(len(active_points)) # Specify the default collision detection region as subduction zones default_collision = reconstruction._DefaultCollision( feature_specific_collision_parameters=[ ( pygplates.FeatureType.gpml_subduction_zone, self.subduction_collision_parameters, ) ] ) # In addition to the default subduction detection, also detect continental collisions # Use the input continent mask if it is provided. if self.continent_mask_filename is not None: collision_spec = reconstruction._ContinentCollision( # This filename string should not have a time formatted into it - this is # taken care of later. self.continent_mask_filename, default_collision, verbose=False, ) else: # If a continent mask is not provided, use the ones made. mask_basename = r"continent_mask_{}Ma.nc" if self.file_collection is not None: mask_basename = str(self.file_collection) + "_" + mask_basename mask_template = os.path.join(self.save_directory, mask_basename) collision_spec = reconstruction._ContinentCollision( mask_template, default_collision, verbose=False, ) # Call the reconstruct by topologies object topology_reconstruction = reconstruction._ReconstructByTopologies( self.rotation_model, self.topology_features, self._max_time, self.min_time, self.ridge_time_step, active_points, point_begin_times=appearance_time, detect_collisions=collision_spec, ) # Initialise the reconstruction. topology_reconstruction.begin_reconstruction() # Loop over the reconstruction times until the end of the reconstruction time span, or until # all points have entered their valid time range *and* either exited their time range or # have been deactivated (subducted forward in time or consumed by MOR backward in time). reconstruction_data = [] while True: print( "Reconstruct by topologies: working on time {:0.2f} Ma".format( topology_reconstruction.get_current_time() ) ) # NOTE: # topology_reconstruction.get_active_current_points() and topology_reconstruction.get_all_current_points() # are different. The former is a subset of the latter, and it represents all points at the timestep that # have not collided with a continental or subduction boundary. The remainders in the latter are inactive # (NoneType) points, which represent the collided points. # We need to access active point data from topology_reconstruction.get_all_current_points() because it has # the same length as the list of all initial ocean points and MOR seed points that have ever emerged from # spreading ridge topologies through `max_time` to `min_time`. Therefore, it protects the time and space # order in which all MOR points through time were seeded by pyGPlates. At any given timestep, not all these # points will be active, but their indices are retained. Thus, z value allocation, point latitudes and # longitudes of active points will be correctly indexed if taking it from # topology_reconstruction.get_all_current_points(). curr_points = topology_reconstruction.get_active_current_points() curr_points_including_inactive = ( topology_reconstruction.get_all_current_points() ) # Collect latitudes and longitudes of currently ACTIVE points in the ocean basin curr_lat_lon_points = [point.to_lat_lon() for point in curr_points] if curr_lat_lon_points: # Get the number of active points at this timestep. num_current_points = len(curr_points) # ndarray to fill with active point lats, lons and zvalues # FOR NOW, the number of gridding input columns is 6: # 0 = longitude # 1 = latitude # 2 = seafloor age # 3 = birth latitude snapshot # 4 = point id # 5 for the default gridding columns above, plus additional zvalues added next total_number_of_columns = 5 + len(self.zval_names) gridding_input_data = np.empty( [num_current_points, total_number_of_columns] ) # Lons and lats are first and second columns of the ndarray respectively gridding_input_data[:, 1], gridding_input_data[:, 0] = zip( *curr_lat_lon_points ) # NOTE: We need a single index to access data from curr_points_including_inactive AND allocate # this data to an ndarray with a number of rows equal to num_current_points. This index will # append +1 after each loop through curr_points_including_inactive. i = 0 # Get indices and points of all points at `time`, both active and inactive (which are NoneType points that # have undergone continental collision or subduction at `time`). for point_index, current_point in enumerate( curr_points_including_inactive ): # Look at all active points (these have not collided with a continent or trench) if current_point is not None: # Seafloor age gridding_input_data[i, 2] = ( appearance_time[point_index] - topology_reconstruction.get_current_time() ) # Birth latitude (snapshot) gridding_input_data[i, 3] = birth_lat[point_index] # Point ID (snapshot) gridding_input_data[i, 4] = point_id[ point_index ] # The ID of a corresponding point from the original list of all MOR-resolved points # GENERAL Z-VALUE ALLOCATION # Z values are 1st index onwards; 0th belongs to the point feature ID (thus +1) for j in range(len(self.zval_names)): # Adjusted index - and we have to add j to 5 to account for lat, lon, age, birth lat and point ID, adjusted_index = 5 + j # Spreading rate would be first # Access current zval from the master list of all zvalues for all points that ever existed in time_array gridding_input_data[i, adjusted_index] = zvalues[ point_index, j ] # Go to the next active point i += 1 gridding_input_dictionary = {} for i in list(range(total_number_of_columns)): gridding_input_dictionary[self.total_column_headers[i]] = [ list(j) for j in zip(*gridding_input_data) ][i] data_to_store = [ gridding_input_dictionary[i] for i in gridding_input_dictionary ] gridding_input_basename = "gridding_input_{:0.1f}Ma".format( topology_reconstruction.get_current_time() ) if self.file_collection is not None: gridding_input_basename = "{}_{}".format( self.file_collection, gridding_input_basename, ) gridding_input_filename = os.path.join( self.save_directory, gridding_input_basename ) np.savez_compressed(gridding_input_filename, *data_to_store) if not topology_reconstruction.reconstruct_to_next_time(): break print( "Reconstruction done for {}!".format( topology_reconstruction.get_current_time() ) ) # return reconstruction_data def lat_lon_z_to_netCDF( self, zval_name, time_arr=None, unmasked=False, nprocs=1, ): """Produce a netCDF4 grid of a z-value identified by its `zval_name` for a given time range in `time_arr`. Seafloor age can be gridded by passing `zval_name` as `SEAFLOOR_AGE`, and spreading rate can be gridded with `SPREADING_RATE`. Saves all grids to compressed netCDF format in the attributed directory. Grids can be read into ndarray format using `gplately.grids.read_netcdf_grid()`. Parameters ---------- zval_name : str A string identifiers for a column in the ReconstructByTopologies gridding input files. time_arr : list of float, default None A time range to turn lons, lats and z-values into netCDF4 grids. If not provided, `time_arr` defaults to the full `time_array` provided to `SeafloorGrids`. unmasked : bool, default False Save unmasked grids, in addition to masked versions. nprocs : int, defaullt 1 Number of processes to use for certain operations (requires joblib). Passed to `joblib.Parallel`, so -1 means all available processes. """ parallel = None nprocs = int(nprocs) if nprocs != 1: try: from joblib import Parallel parallel = Parallel(nprocs) except ImportError: warnings.warn( "Could not import joblib; falling back to serial execution" ) # User can put any time array within SeafloorGrid bounds, but if none # is provided, it defaults to the attributed time array if time_arr is None: time_arr = self.time_array if parallel is None: for time in time_arr: _lat_lon_z_to_netCDF_time( time=time, zval_name=zval_name, file_collection=self.file_collection, save_directory=self.save_directory, total_column_headers=self.total_column_headers, extent=self.extent, resX=self.spacingX, resY=self.spacingY, unmasked=unmasked, continent_mask_filename=self.continent_mask_filename, ) else: from joblib import delayed parallel( delayed(_lat_lon_z_to_netCDF_time)( time=time, zval_name=zval_name, file_collection=self.file_collection, save_directory=self.save_directory, total_column_headers=self.total_column_headers, extent=self.extent, resX=self.spacingX, resY=self.spacingY, unmasked=unmasked, continent_mask_filename=self.continent_mask_filename, ) for time in time_arr )Instance variables
var PlotTopologiesTime-
Expand source code
@property def PlotTopologiesTime(self): return self._PlotTopologies_object.time var max_time-
The reconstruction time.
Expand source code
@property def max_time(self): """The reconstruction time.""" return self._max_time
Methods
def build_all_MOR_seedpoints(self)-
Resolve mid-ocean ridges for all times between
min_timeandmax_time, divide them into points that make up their shared sub-segments. Rotate these points to the left and right of the ridge using their stage rotation so that they spread from the ridge.Z-value allocation to each point is done here. In future, a function (like the spreading rate function) to calculate general z-data will be an input parameter.
Notes
If MOR seed point building is interrupted, progress is safeguarded as long as
resume_from_checkpointsis set toTrue.This assumes that points spread from ridges symmetrically, with the exception of large ridge jumps at successive timesteps. Therefore, z-values allocated to ridge-emerging points will appear symmetrical until changes in spreading ridge geometries create asymmetries.
In future, this will have a checkpoint save feature so that execution (which occurs during preparation for ReconstructByTopologies and can take several hours) can be safeguarded against run interruptions.
Parameters
time:float- The time at which to resolve ridge points and stage-rotate them off the ridge.
Returns
mor_point_features:FeatureCollection- All ridge seed points that have emerged from all ridge topologies at
time. These points have spread by being slightly rotated away from ridge locations attime.
References
get_mid_ocean_ridge_seedpoints() has been adapted from https://github.com/siwill22/agegrid-0.1/blob/master/automatic_age_grid_seeding.py#L117.
Expand source code
def build_all_MOR_seedpoints(self): """Resolve mid-ocean ridges for all times between `min_time` and `max_time`, divide them into points that make up their shared sub-segments. Rotate these points to the left and right of the ridge using their stage rotation so that they spread from the ridge. Z-value allocation to each point is done here. In future, a function (like the spreading rate function) to calculate general z-data will be an input parameter. Notes ----- If MOR seed point building is interrupted, progress is safeguarded as long as `resume_from_checkpoints` is set to `True`. This assumes that points spread from ridges symmetrically, with the exception of large ridge jumps at successive timesteps. Therefore, z-values allocated to ridge-emerging points will appear symmetrical until changes in spreading ridge geometries create asymmetries. In future, this will have a checkpoint save feature so that execution (which occurs during preparation for ReconstructByTopologies and can take several hours) can be safeguarded against run interruptions. Parameters ---------- time : float The time at which to resolve ridge points and stage-rotate them off the ridge. Returns ------- mor_point_features : FeatureCollection All ridge seed points that have emerged from all ridge topologies at `time`. These points have spread by being slightly rotated away from ridge locations at `time`. References ---------- get_mid_ocean_ridge_seedpoints() has been adapted from https://github.com/siwill22/agegrid-0.1/blob/master/automatic_age_grid_seeding.py#L117. """ # If we mustn't overwrite existing files in the `save_directory`, check the status of MOR seeding # to know where to start/continue seeding if self.resume_from_checkpoints: # Check the last MOR seedpoint gpmlz file that was built checkpointed_MOR_seedpoints = [ s.split("/")[-1] for s in glob.glob(self.save_directory + "/" + "*MOR_plus_one_points*") ] try: # -2 as an index accesses the age (float type), safeguards against identifying numbers in the SeafloorGrid.file_collection string last_seed_time = np.sort( [ float(re.findall(r"\d+", s)[-2]) for s in checkpointed_MOR_seedpoints ] )[0] # If none were built yet except: last_seed_time = "nil" # If MOR seeding has not started, start it from the top if last_seed_time == "nil": time_array = self.time_array # If the last seed time it could identify is outside the time bounds of the current instance of SeafloorGrid, start # from the top (this may happen if we use the same save directory for grids for a new set of times) elif last_seed_time not in self.time_array: time_array = self.time_array # If seeding was done to the min_time, we are finished elif last_seed_time == self.min_time: return # If seeding to `min_time` has been interrupted, resume it at last_masked_time. else: time_array = np.arange( last_seed_time, self.min_time - 0.1, -self.ridge_time_step ) # If we must overwrite all files in `save_directory`, start from `max_time`. else: time_array = self.time_array # Build all continental masks and spreading ridge points (with z values) self._get_mid_ocean_ridge_seedpoints(time_array) return def build_all_continental_masks(self)-
Create a continental mask to define the ocean basin for all times between
min_timeandmax_time. as well as to use as continental collision boundaries inReconstructByTopologies.Notes
Continental masking progress is safeguarded if ever masking is interrupted, provided that
resume_from_checkpointsis set toTrue.If
ReconstructByTopologiesidentifies a continental collision between oceanic points and the boundaries of this continental mask attime, those points are deleted attime.The continental mask is also saved to "/continent_mask_{}Ma.nc" as a compressed netCDF4 file if a
save_directoryis passed. Otherwise, the final grid is returned as a NumPy ndarray object.Returns
all_continental_masks:listofndarray- A masked grid per timestep in
time_arraywith 1=continental point, and 0=ocean point, for all points on the full global icosahedral mesh.
Expand source code
def build_all_continental_masks(self): """Create a continental mask to define the ocean basin for all times between `min_time` and `max_time`. as well as to use as continental collision boundaries in `ReconstructByTopologies`. Notes ----- Continental masking progress is safeguarded if ever masking is interrupted, provided that `resume_from_checkpoints` is set to `True`. If `ReconstructByTopologies` identifies a continental collision between oceanic points and the boundaries of this continental mask at `time`, those points are deleted at `time`. The continental mask is also saved to "/continent_mask_{}Ma.nc" as a compressed netCDF4 file if a `save_directory` is passed. Otherwise, the final grid is returned as a NumPy ndarray object. Returns ------- all_continental_masks : list of ndarray A masked grid per timestep in `time_array` with 1=continental point, and 0=ocean point, for all points on the full global icosahedral mesh. """ # If we mustn't overwrite existing files in the `save_directory`, check the status # of continental masking to know where to start/continue masking if self.resume_from_checkpoints: # Check the last continental mask that could be built checkpointed_continental_masks = [ s.split("/")[-1] for s in glob.glob(self.save_directory + "/" + "*continent_mask*") ] try: # -2 as an index accesses the age (float type), safeguards against identifying numbers in the SeafloorGrid.file_collection string last_masked_time = np.sort( [ float(re.findall(r"\d+", s)[-2]) for s in checkpointed_continental_masks ] )[0] # If none were built yet except: last_masked_time = "nil" # If masking has not started, start it from the top if last_masked_time == "nil": time_array = self.time_array # If the last seed time it could identify is outside the time bounds of the current instance of SeafloorGrid, start # from the top (this may happen if we use the same save directory for grids for a new set of times) elif last_masked_time not in self.time_array: time_array = self.time_array # If masking was done to the min_time, we are finished elif last_masked_time == self.min_time: return # If masking to `min_time` has been interrupted, resume it at last_masked_time. else: time_array = np.arange( last_masked_time, self.min_time - 0.1, -self.ridge_time_step ) # If we must overwrite all files in `save_directory`, start from `max_time`. else: time_array = self.time_array # Build all continental masks and spreading ridge points (with z values) self._create_continental_mask(time_array) return def create_initial_ocean_seed_points(self)-
Create the initial ocean basin seed point domain (at
max_timeonly) using Stripy's icosahedral triangulation with the specifiedself.refinement_levels.The ocean mesh starts off as a global-spanning Stripy icosahedral mesh.
create_initial_ocean_seed_pointspasses the automatically-resolved-to-current-time continental polygons from thePlotTopologies_object'scontinentsattribute (which can be from a COB terrane file or a continental polygon file) into Plate Tectonic Tools' point-in-polygon routine. It identifies ocean basin points that lie: * outside the polygons (for the ocean basin point domain) * inside the polygons (for the continental mask)Points from the mesh outside the continental polygons make up the ocean basin seed point mesh. The masked mesh is outputted as a compressed GPML (GPMLZ) file with the filename: "ocean_basin_seed_points_{}Ma.gpmlz" if a
save_directoryis passed. Otherwise, the mesh is returned as a pyGPlates FeatureCollection object.Notes
This point mesh represents ocean basin seafloor that was produced before
SeafloorGrid.max_time, and thus has unknown properties like valid time and spreading rate. As time passes, the plate reconstruction model sees points emerging from MORs. These new points spread to occupy the ocean basins, moving the initial filler points closer to subduction zones and continental polygons with which they can collide. If a collision is detected byPlateReconstructionsReconstructByTopologiesobject, these points are deleted.Ideally, if a reconstruction tree spans a large time range, all initial mesh points would collide with a continent or be subducted, leaving behind a mesh of well-defined MOR-emerged ocean basin points that data can be attributed to. However, some of these initial points situated close to contiental boundaries are retained through time - these form point artefacts with anomalously high ages. Even deep-time plate models (e.g. 1 Ga) will have these artefacts - removing them would require more detail to be added to the reconstruction model.
Returns
ocean_basin_point_mesh:FeatureCollectionofpygplates.MultiPointOnSphere- A feature collection of point objects on the ocean basin.
Expand source code
def create_initial_ocean_seed_points(self): """Create the initial ocean basin seed point domain (at `max_time` only) using Stripy's icosahedral triangulation with the specified `self.refinement_levels`. The ocean mesh starts off as a global-spanning Stripy icosahedral mesh. `create_initial_ocean_seed_points` passes the automatically-resolved-to-current-time continental polygons from the `PlotTopologies_object`'s `continents` attribute (which can be from a COB terrane file or a continental polygon file) into Plate Tectonic Tools' point-in-polygon routine. It identifies ocean basin points that lie: * outside the polygons (for the ocean basin point domain) * inside the polygons (for the continental mask) Points from the mesh outside the continental polygons make up the ocean basin seed point mesh. The masked mesh is outputted as a compressed GPML (GPMLZ) file with the filename: "ocean_basin_seed_points_{}Ma.gpmlz" if a `save_directory` is passed. Otherwise, the mesh is returned as a pyGPlates FeatureCollection object. Notes ----- This point mesh represents ocean basin seafloor that was produced before `SeafloorGrid.max_time`, and thus has unknown properties like valid time and spreading rate. As time passes, the plate reconstruction model sees points emerging from MORs. These new points spread to occupy the ocean basins, moving the initial filler points closer to subduction zones and continental polygons with which they can collide. If a collision is detected by `PlateReconstruction`s `ReconstructByTopologies` object, these points are deleted. Ideally, if a reconstruction tree spans a large time range, **all** initial mesh points would collide with a continent or be subducted, leaving behind a mesh of well-defined MOR-emerged ocean basin points that data can be attributed to. However, some of these initial points situated close to contiental boundaries are retained through time - these form point artefacts with anomalously high ages. Even deep-time plate models (e.g. 1 Ga) will have these artefacts - removing them would require more detail to be added to the reconstruction model. Returns ------- ocean_basin_point_mesh : pygplates.FeatureCollection of pygplates.MultiPointOnSphere A feature collection of point objects on the ocean basin. """ if self.continent_mask_filename is None: # Ensure COB terranes at max time have reconstruction IDs and valid times COB_polygons = ensure_polygon_geometry( self._PlotTopologies_object.continents, self.rotation_model, self._max_time, ) # zval is a binary array encoding whether a point # coordinate is within a COB terrane polygon or not. # Use the icosahedral mesh MultiPointOnSphere attribute _, ocean_basin_point_mesh, zvals = point_in_polygon_routine( self.icosahedral_multi_point, COB_polygons ) # Plates to partition with plate_partitioner = pygplates.PlatePartitioner( COB_polygons, self.rotation_model, ) # Plate partition the ocean basin points meshnode_feature = pygplates.Feature( pygplates.FeatureType.create_from_qualified_string("gpml:MeshNode") ) meshnode_feature.set_geometry( ocean_basin_point_mesh # multi_point ) ocean_basin_meshnode = pygplates.FeatureCollection(meshnode_feature) paleogeography = plate_partitioner.partition_features( ocean_basin_meshnode, partition_return=pygplates.PartitionReturn.separate_partitioned_and_unpartitioned, properties_to_copy=[pygplates.PropertyName.gpml_shapefile_attributes], ) ocean_points = paleogeography[1] # Separate those inside polygons continent_points = paleogeography[0] # Separate those outside polygons # If a set of continent masks was passed, we can use max_time's continental # mask to build the initial profile of seafloor age. else: max_time_cont_mask = grids.Raster( self.continent_mask_filename.format(self._max_time) ) # If the input grid is at 0.5 degree uniform spacing, then the input # grid is 7x more populated than a 6-level stripy icosahedral mesh and # using this resolution for the initial ocean mesh will dramatically slow down # reconstruction by topologies. # Scale down the resolution based on the input mask resolution # (percentage was found in __init__.) max_time_cont_mask.resize( int(max_time_cont_mask.shape[0] * self.percentage), int(max_time_cont_mask.shape[1] * self.percentage), inplace=True, ) lat = np.linspace(-90, 90, max_time_cont_mask.shape[0]) lon = np.linspace(-180, 180, max_time_cont_mask.shape[1]) llon, llat = np.meshgrid(lon, lat) mask_inds = np.where(max_time_cont_mask.data.flatten() == 0) mask_vals = max_time_cont_mask.data.flatten() mask_lon = llon.flatten()[mask_inds] mask_lat = llat.flatten()[mask_inds] ocean_pt_feature = pygplates.Feature() ocean_pt_feature.set_geometry( pygplates.MultiPointOnSphere(zip(mask_lat, mask_lon)) ) ocean_points = [ocean_pt_feature] # Now that we have ocean points... # Determine age of ocean basin points using their proximity to MOR features # and an assumed globally-uniform ocean basin mean spreading rate. # We need resolved topologies at the `max_time` to pass into the proximity # function resolved_topologies = [] shared_boundary_sections = [] pygplates.resolve_topologies( self.topology_features, self.rotation_model, resolved_topologies, self._max_time, shared_boundary_sections, ) pX, pY, pZ = tools.find_distance_to_nearest_ridge( resolved_topologies, shared_boundary_sections, ocean_points, ) # Divide spreading rate by 2 to use half the mean spreading rate pAge = np.array(pZ) / (self.initial_ocean_mean_spreading_rate / 2.0) initial_ocean_point_features = [] initial_ocean_multipoints = [] for point in zip(pX, pY, pAge): point_feature = pygplates.Feature() point_feature.set_geometry(pygplates.PointOnSphere(point[1], point[0])) # Add 'time' to the age at the time of computation, to get the valid time in Ma point_feature.set_valid_time(point[2] + self._max_time, -1) # For now: custom zvals are added as shapefile attributes - will attempt pandas data frames # point_feature = set_shapefile_attribute(point_feature, self.initial_ocean_mean_spreading_rate, "SPREADING_RATE") # Seems like static data initial_ocean_point_features.append(point_feature) initial_ocean_multipoints.append(point_feature.get_geometry()) # print(initial_ocean_point_features) multi_point_feature = pygplates.MultiPointOnSphere(initial_ocean_multipoints) basename = "ocean_basin_seed_points_{}_RLs_{}Ma.gpmlz".format( self.refinement_levels, self._max_time, ) if self.file_collection is not None: basename = "{}_{}".format(self.file_collection, basename) output_filename = os.path.join(self.save_directory, basename) initial_ocean_feature_collection = pygplates.FeatureCollection( initial_ocean_point_features ) initial_ocean_feature_collection.write(output_filename) # Collect all point feature data into a pandas dataframe self._collect_point_data_in_dataframe( initial_ocean_feature_collection, np.array( [self.initial_ocean_mean_spreading_rate] * len(pX) ), # for now, spreading rate is one zvalue for initial ocean points. will other zvalues need to have a generalised workflow? self._max_time, ) return ( pygplates.FeatureCollection(initial_ocean_point_features), multi_point_feature, ) def lat_lon_z_to_netCDF(self, zval_name, time_arr=None, unmasked=False, nprocs=1)-
Produce a netCDF4 grid of a z-value identified by its
zval_namefor a given time range intime_arr.Seafloor age can be gridded by passing
zval_nameasSEAFLOOR_AGE, and spreading rate can be gridded withSPREADING_RATE.Saves all grids to compressed netCDF format in the attributed directory. Grids can be read into ndarray format using
read_netcdf_grid().Parameters
zval_name:str- A string identifiers for a column in the ReconstructByTopologies gridding input files.
time_arr:listoffloat, defaultNone- A time range to turn lons, lats and z-values into netCDF4 grids. If not provided,
time_arrdefaults to the fulltime_arrayprovided toSeafloorGrids. unmasked:bool, defaultFalse- Save unmasked grids, in addition to masked versions.
nprocs:int, defaullt 1- Number of processes to use for certain operations (requires joblib).
Passed to
joblib.Parallel, so -1 means all available processes.
Expand source code
def lat_lon_z_to_netCDF( self, zval_name, time_arr=None, unmasked=False, nprocs=1, ): """Produce a netCDF4 grid of a z-value identified by its `zval_name` for a given time range in `time_arr`. Seafloor age can be gridded by passing `zval_name` as `SEAFLOOR_AGE`, and spreading rate can be gridded with `SPREADING_RATE`. Saves all grids to compressed netCDF format in the attributed directory. Grids can be read into ndarray format using `gplately.grids.read_netcdf_grid()`. Parameters ---------- zval_name : str A string identifiers for a column in the ReconstructByTopologies gridding input files. time_arr : list of float, default None A time range to turn lons, lats and z-values into netCDF4 grids. If not provided, `time_arr` defaults to the full `time_array` provided to `SeafloorGrids`. unmasked : bool, default False Save unmasked grids, in addition to masked versions. nprocs : int, defaullt 1 Number of processes to use for certain operations (requires joblib). Passed to `joblib.Parallel`, so -1 means all available processes. """ parallel = None nprocs = int(nprocs) if nprocs != 1: try: from joblib import Parallel parallel = Parallel(nprocs) except ImportError: warnings.warn( "Could not import joblib; falling back to serial execution" ) # User can put any time array within SeafloorGrid bounds, but if none # is provided, it defaults to the attributed time array if time_arr is None: time_arr = self.time_array if parallel is None: for time in time_arr: _lat_lon_z_to_netCDF_time( time=time, zval_name=zval_name, file_collection=self.file_collection, save_directory=self.save_directory, total_column_headers=self.total_column_headers, extent=self.extent, resX=self.spacingX, resY=self.spacingY, unmasked=unmasked, continent_mask_filename=self.continent_mask_filename, ) else: from joblib import delayed parallel( delayed(_lat_lon_z_to_netCDF_time)( time=time, zval_name=zval_name, file_collection=self.file_collection, save_directory=self.save_directory, total_column_headers=self.total_column_headers, extent=self.extent, resX=self.spacingX, resY=self.spacingY, unmasked=unmasked, continent_mask_filename=self.continent_mask_filename, ) for time in time_arr ) def prepare_for_reconstruction_by_topologies(self)-
Prepare three main auxiliary files for seafloor data gridding: * Initial ocean seed points (at
max_time) * Continental masks (frommax_timetomin_time) * MOR points (frommax_timetomin_time)Returns lists of all attributes for the initial ocean point mesh and all ridge points for all times in the reconstruction time array.
Expand source code
def prepare_for_reconstruction_by_topologies(self): """Prepare three main auxiliary files for seafloor data gridding: * Initial ocean seed points (at `max_time`) * Continental masks (from `max_time` to `min_time`) * MOR points (from `max_time` to `min_time`) Returns lists of all attributes for the initial ocean point mesh and all ridge points for all times in the reconstruction time array. """ # INITIAL OCEAN SEED POINT MESH ---------------------------------------------------- ( initial_ocean_seed_points, initial_ocean_seed_points_mp, ) = self.create_initial_ocean_seed_points() print("Finished building initial_ocean_seed_points!") # MOR SEED POINTS AND CONTINENTAL MASKS -------------------------------------------- # The start time for seeding is controlled by whether the overwrite_existing_gridding_inputs # parameter is set to `True` (in which case the start time is `max_time`). If it is `False` # and; # - a run of seeding and continental masking was interrupted, and ridge points were # checkpointed at n Ma, seeding resumes at n-1 Ma until `min_time` or another interruption # occurs; # - seeding was completed but the subsequent gridding input creation was interrupted, # seeding is assumed completed and skipped. The workflow automatically proceeds to re-gridding. if self.continent_mask_filename is None: self.build_all_continental_masks() else: print( "Continent masks passed to SeafloorGrid - skipping continental mask generation!" ) self.build_all_MOR_seedpoints() # ALL-TIME POINTS ----------------------------------------------------- # Extract all feature attributes for all reconstruction times into lists active_points = [] appearance_time = [] birth_lat = [] # latitude_of_crust_formation prev_lat = [] prev_lon = [] # Extract point feature attributes from MOR seed points all_mor_features = [] zvalues = np.empty((0, len(self.zval_names))) for time in self.time_array: # If we're at the maximum time, start preparing points from the initial ocean mesh # as well as their z values if time == self._max_time: for feature in initial_ocean_seed_points: active_points.append(feature.get_geometry()) appearance_time.append(feature.get_valid_time()[0]) birth_lat.append(feature.get_geometry().to_lat_lon_list()[0][0]) prev_lat.append(feature.get_geometry().to_lat_lon_list()[0][0]) prev_lon.append(feature.get_geometry().to_lat_lon_list()[0][1]) curr_zvalues = self._extract_zvalues_from_npz_to_ndarray( initial_ocean_seed_points, time ) zvalues = np.concatenate((zvalues, curr_zvalues), axis=0) # Otherwise, we'd be preparing MOR points and their z values else: # GPMLZ file of MOR seedpoints basename = "MOR_plus_one_points_{:0.2f}.gpmlz".format(time) if self.file_collection is not None: basename = "{}_{}".format(self.file_collection, basename) filename = os.path.join(self.save_directory, basename) features = pygplates.FeatureCollection(filename) for feature in features: if feature.get_valid_time()[0] < self.time_array[0]: active_points.append(feature.get_geometry()) appearance_time.append(feature.get_valid_time()[0]) birth_lat.append(feature.get_geometry().to_lat_lon_list()[0][0]) prev_lat.append(feature.get_geometry().to_lat_lon_list()[0][0]) prev_lon.append(feature.get_geometry().to_lat_lon_list()[0][1]) # COLLECT NDARRAY OF ALL ZVALUES IN THIS TIMESTEP ------------------ curr_zvalues = self._extract_zvalues_from_npz_to_ndarray(features, time) zvalues = np.concatenate((zvalues, curr_zvalues), axis=0) return active_points, appearance_time, birth_lat, prev_lat, prev_lon, zvalues def reconstruct_by_topologies(self)-
Obtain all active ocean seed points at
time- these are points that have not been consumed at subduction zones or have not collided with continental polygons.All active points' latitudes, longitues, seafloor ages, spreading rates and all other general z-values are saved to a gridding input file (.npz).
Expand source code
def reconstruct_by_topologies(self): """Obtain all active ocean seed points at `time` - these are points that have not been consumed at subduction zones or have not collided with continental polygons. All active points' latitudes, longitues, seafloor ages, spreading rates and all other general z-values are saved to a gridding input file (.npz). """ print("Preparing all initial files...") # Obtain all info from the ocean seed points and all MOR points through time, store in # arrays ( active_points, appearance_time, birth_lat, prev_lat, prev_lon, zvalues, ) = self.prepare_for_reconstruction_by_topologies() #### Begin reconstruction by topology process: # Indices for all points (`active_points`) that have existed from `max_time` to `min_time`. point_id = range(len(active_points)) # Specify the default collision detection region as subduction zones default_collision = reconstruction._DefaultCollision( feature_specific_collision_parameters=[ ( pygplates.FeatureType.gpml_subduction_zone, self.subduction_collision_parameters, ) ] ) # In addition to the default subduction detection, also detect continental collisions # Use the input continent mask if it is provided. if self.continent_mask_filename is not None: collision_spec = reconstruction._ContinentCollision( # This filename string should not have a time formatted into it - this is # taken care of later. self.continent_mask_filename, default_collision, verbose=False, ) else: # If a continent mask is not provided, use the ones made. mask_basename = r"continent_mask_{}Ma.nc" if self.file_collection is not None: mask_basename = str(self.file_collection) + "_" + mask_basename mask_template = os.path.join(self.save_directory, mask_basename) collision_spec = reconstruction._ContinentCollision( mask_template, default_collision, verbose=False, ) # Call the reconstruct by topologies object topology_reconstruction = reconstruction._ReconstructByTopologies( self.rotation_model, self.topology_features, self._max_time, self.min_time, self.ridge_time_step, active_points, point_begin_times=appearance_time, detect_collisions=collision_spec, ) # Initialise the reconstruction. topology_reconstruction.begin_reconstruction() # Loop over the reconstruction times until the end of the reconstruction time span, or until # all points have entered their valid time range *and* either exited their time range or # have been deactivated (subducted forward in time or consumed by MOR backward in time). reconstruction_data = [] while True: print( "Reconstruct by topologies: working on time {:0.2f} Ma".format( topology_reconstruction.get_current_time() ) ) # NOTE: # topology_reconstruction.get_active_current_points() and topology_reconstruction.get_all_current_points() # are different. The former is a subset of the latter, and it represents all points at the timestep that # have not collided with a continental or subduction boundary. The remainders in the latter are inactive # (NoneType) points, which represent the collided points. # We need to access active point data from topology_reconstruction.get_all_current_points() because it has # the same length as the list of all initial ocean points and MOR seed points that have ever emerged from # spreading ridge topologies through `max_time` to `min_time`. Therefore, it protects the time and space # order in which all MOR points through time were seeded by pyGPlates. At any given timestep, not all these # points will be active, but their indices are retained. Thus, z value allocation, point latitudes and # longitudes of active points will be correctly indexed if taking it from # topology_reconstruction.get_all_current_points(). curr_points = topology_reconstruction.get_active_current_points() curr_points_including_inactive = ( topology_reconstruction.get_all_current_points() ) # Collect latitudes and longitudes of currently ACTIVE points in the ocean basin curr_lat_lon_points = [point.to_lat_lon() for point in curr_points] if curr_lat_lon_points: # Get the number of active points at this timestep. num_current_points = len(curr_points) # ndarray to fill with active point lats, lons and zvalues # FOR NOW, the number of gridding input columns is 6: # 0 = longitude # 1 = latitude # 2 = seafloor age # 3 = birth latitude snapshot # 4 = point id # 5 for the default gridding columns above, plus additional zvalues added next total_number_of_columns = 5 + len(self.zval_names) gridding_input_data = np.empty( [num_current_points, total_number_of_columns] ) # Lons and lats are first and second columns of the ndarray respectively gridding_input_data[:, 1], gridding_input_data[:, 0] = zip( *curr_lat_lon_points ) # NOTE: We need a single index to access data from curr_points_including_inactive AND allocate # this data to an ndarray with a number of rows equal to num_current_points. This index will # append +1 after each loop through curr_points_including_inactive. i = 0 # Get indices and points of all points at `time`, both active and inactive (which are NoneType points that # have undergone continental collision or subduction at `time`). for point_index, current_point in enumerate( curr_points_including_inactive ): # Look at all active points (these have not collided with a continent or trench) if current_point is not None: # Seafloor age gridding_input_data[i, 2] = ( appearance_time[point_index] - topology_reconstruction.get_current_time() ) # Birth latitude (snapshot) gridding_input_data[i, 3] = birth_lat[point_index] # Point ID (snapshot) gridding_input_data[i, 4] = point_id[ point_index ] # The ID of a corresponding point from the original list of all MOR-resolved points # GENERAL Z-VALUE ALLOCATION # Z values are 1st index onwards; 0th belongs to the point feature ID (thus +1) for j in range(len(self.zval_names)): # Adjusted index - and we have to add j to 5 to account for lat, lon, age, birth lat and point ID, adjusted_index = 5 + j # Spreading rate would be first # Access current zval from the master list of all zvalues for all points that ever existed in time_array gridding_input_data[i, adjusted_index] = zvalues[ point_index, j ] # Go to the next active point i += 1 gridding_input_dictionary = {} for i in list(range(total_number_of_columns)): gridding_input_dictionary[self.total_column_headers[i]] = [ list(j) for j in zip(*gridding_input_data) ][i] data_to_store = [ gridding_input_dictionary[i] for i in gridding_input_dictionary ] gridding_input_basename = "gridding_input_{:0.1f}Ma".format( topology_reconstruction.get_current_time() ) if self.file_collection is not None: gridding_input_basename = "{}_{}".format( self.file_collection, gridding_input_basename, ) gridding_input_filename = os.path.join( self.save_directory, gridding_input_basename ) np.savez_compressed(gridding_input_filename, *data_to_store) if not topology_reconstruction.reconstruct_to_next_time(): break print( "Reconstruction done for {}!".format( topology_reconstruction.get_current_time() ) ) # return reconstruction_data def update_time(self, max_time)-
Expand source code
def update_time(self, max_time): self._max_time = float(max_time) self._PlotTopologies_object.time = float(max_time)